Open Access | Research
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Dysregulation of Nrf2/ARE pathway and mitochondrial DNA damage in aging and Alzheimer’s-associated cognitive decline
* Corresponding author: Polina I. Babenkova
Mailing address: Department of Genetics, Cytology and Bioengineering, Voronezh State University, Voronezh, Russia.
Email: ms.babenkova@gmail.com
Received: 02 July 2025 / Revised: 01 August 2025 / Accepted: 04 September 2025 / Published: 30 December 2025
DOI: 10.31491/APT.2025.09.184
Abstract
Background: Mitochondrial dysfunction represents a critical link between physiological aging and Alzheimer’s disease (AD), though the specific mechanisms driving pathological cognitive decline remain unclear. In this study, we investigated how Nrf2/ARE pathway suppression and mitochondrial DNA (mtDNA) damage contribute to accelerated memory impairment in APP/PS1 mice compared to age-matched wild-type (C57BL/6) controls.
Materials and methods: The Barnes test was used to assess cognitive function. We also analyzed the level of oxidative damage to mitochondrial DNA (mtDNA) in various areas of the brain, including the cerebellum and midbrain. We evaluated the activity of the Nrf2/ARE cytoprotective pathway, as well as the expression of antioxidant genes involved in H2O2 utilization and genes associated with DNA repair. Additionally, we investigated the level of the pro-inflammatory cytokine TNF-α and changes in the composition of the intestinal microbiota.
Results: Cognitive impairments were observed in transgenic mice at 4 and 8 months of age, as evidenced by an increase in the time required to find the exit. Transgenic mice showed a significant accumulation of oxidative damage to mitochondrial DNA (mtDNA) in certain areas of the brain, primarily in the cerebellum and partially in the midbrain. Compared to C57BL/6 mice, APP/PS1 mice showed reduced expression of antioxidant genes involved in H2O2 utilization, as well as genes associated with DNA repair. These mitochondrial dysfunctions may contribute to cerebellar dysfunction. We found increased expression of the pro-inflammatory cytokine TNFα, as well as some signs of changes in the composition of the gut microbiota.
Conclusion: Our findings identify mitochondrial maintenance, particularly via the Nrf2/ARE pathway, as a promising target for interventions aimed at mitigating pathological aging while preserving normal cognitive function.
Keywords
Alzheimer’s disease, cognitive impairments, Barnes maze, APP/PS1 mouse strain, Nrf2/ARE signal pathway
Introduction
Aging is a multifactorial process affecting all levels of biological organization from molecular changes to systemic disorders. One of the key aspects of aging is the gradual accumulation of damage in cells and tissues caused by oxidative stress, impaired reparative mechanisms and dysfunction of protein homeostasis [1]. These changes create the preconditions for the development of age-associated diseases, among which neurodegenerative pathologies, in particular Alzheimer’s disease (AD), occupy a special place [2]. More than 90% of AD cases are sporadic and usually manifest in old age after 65 years [3]. It is believed that the primary cause of AD are aggregation and accumulation of amyloid-β peptide (Aβ) in the brain, which is produced by proteolytic cleavage of amyloid precursor protein (APP) [4]. Proteolytic cleavage of APP is catalyzed by beta-secretase and gamma-secretase. Presenilin (PSEN1/2) is a subcomponent of gamma-secretase [5]. Therefore, the main cause of familial AD is inheritance of mutant alleles in the APP and PSEN1/2 genes [3].
Dementia, which according to some studies is a manifestation of, specifically, AD in animals is found, for example, in dogs, [6, 7] transgenic mouse models expressing human mutant APP and PSEN1/2 genes have been created [8]. These mice exhibit Aβ accumulation in the brain, leading to the development of inflammatory processes and cognitive deterioration [9]. It has been shown that these transgenic mice have mitochondrial dysfunction similar to those observed in patients with AD [10, 11]. However, it has not previously been shown whether transgenic AD model mice are able to accumulate mtDNA damage similar to AD patients [12]. The nuclear DNA of APP/PS1 mice has been shown to accumulate more oxidative damage [13, 14], and mtDNA copy number is reduced due to impaired mitochondrial biogenesis [15, 16], but the effect of genotype on mtDNA damage levels has not been studied.
It should be taken into account that the maintenance of mtDNA integrity is a complex coordinated process that requires correct operation of antioxidant defense systems [17], mechanisms for repair of damaged bases or broken mtDNA strands [18] and elimination systems of old damaged mitochondria via selective mitochondrial autophagy known as mitophagy [19]. These processes are united by the fact that they are regulated at the transcriptional level by the nuclear erythroid 2-related factor 2 (Nrf2, encoded Nfe2l2 gene) / antioxidant response element (ARE) signaling pathway [20-22]. Reduced activity of the Nrf2/ARE signaling pathway is characteristic of AD. AD drugs currently being tested in clinical and preclinical trials are often Nrf2 activators [23]. Moreover, the activity of the Nrf2/ARE signaling pathway is closely related to inflammatory signals in the body [24]. Inflammation, in its turn, can be induced not only by external factors but also by the structural components of the bacteria that constitute the gut microbiome [25]. Although changes in the bacterial composition of the gut microbiome have been repeatedly studied in both AD patients [26-28] and transgenic mice [29-31], there are no studies that link changes in the gut microbiome and inflammation to Nrf2/ARE signaling pathway activity, mtDNA stability and cognitive function in mice.
The aim of this work was to identify cognitive abnormalities in APP/PS1 mice at 4 and 8 months of age and to examine their relationship with the amount of mtDNA damage, the expression of genes encoding proteins involved in antioxidant defense, DNA repair, mitophagy, inflammation and associated with Nrf2/ARE signaling pathway in four brain compartments, and changes in the bacterial composition of the gut microbiome at phylum level.
Materials and methods
Animals and experiment design
In this study, male and female mice of the C57BL/6 and APP/PS1 strains were used. The C57BL/6 mice were obtained from Stolbovaya breeding and nursery laboratory (Research Center for Biomedical Technologies of FMBA; Russia). The APP/PS1 mice (B6C3-Tg(APP695)85Dbo Tg(PSENI)85Dbo strain) were received from Pushchino Nursery for Laboratory Animals (Russia, Pushchino). All experimental procedures including sample sizes were approved by the ethical commission of Voronezh State University (Section of Animal Care and Use, protocol 42-03, 94 of October 8, 2020) in accordance with the principles of the 3R and the requirements of Directive 2010/63/EU of the European Parliament and the Council of the European Union on the protection of animals used for scientific purposes.
All animals were maintained under controlled conditions of 12-hour light/12-hour dark cycle, at 25 °C, received unrestricted water and a standard laboratory diet (Ssniff-Spezialdiäten GmbH, Germany). 4-month-old non-transgenic (C57BL/6 strain; n = 7) and transgenic mice (APP/PS1 strain; n = 10) were subjected to cognitive function testing using the Barnes maze. At this stage, fecal samples were collected from the mice for subsequent DNA extraction to study the bacterial composition of the gut microbiome. The mice were not euthanized at this stage and continued to be housed in the vivarium until they reached 8 months of age. Behavioral experiments and feces collection were also conducted when the mice reached 8 months of age (one C57BL/6 mouse died, resulting in a final C57BL/6 strain sample size of n = 6). Subsequently, the mice were euthanized, and various brain compartments such as the cerebral cortex, hippocampus, midbrain, and cerebellum were isolated for DNA extraction to study mtDNA damage and RNA extraction to study gene expression.
Barnes maze test
To assess the cognitive parameters of the mice, we utilized the Barnes maze and followed the protocol [32]. The Barnes maze test apparatus is an elevated circular plat-form with 40 evenly spaced holes around its perimeter. An escape hole was placed under one hole, while the other 39 holes were left empty. Bright lighting of 1,000 lux and white noise of 40 dB volume were used as stressors. The test consisted of several phases, including adaptation, training, and probe.
During the adaptation phase, lights and sound were turned off. The mouse was placed in the escape hole for 1 minute. Next, the mouse was positioned in the starting chamber, which was placed in the center of the platform for 15 seconds. The allotted time for adaptation lasted 5 minutes. After that, the mouse was placed in the escape hole for 1 minute.
During the training phase, lights and sound were turned on. The mouse was placed in the starting chamber on a specific quadrant. A list of the location of the starting chamber is presented in Table 1. The allotted time to find the escape hole was 3 minutes. After the mouse climbed into the escape hole, the sound and lights were turned off. If the mouse did not climb into the escape hole, it was forcibly placed there and remained inside for 30 seconds. Each mouse was given two attempts per day.
Table 1.
A list of the location of the starting chamber.
| Day | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| First attempt | N | SE | NW | E | N | Сenter of the maze | S | NW | SE | Сenter of the maze |
| Second attempt | E | N | SE | NW | SE | W | S | NW |
During the testing phase on the 6th day, the lights and sound were turned on. The escape hole was closed with the same plug as the other openings. The mouse was placed in the center of the platform in the starting chamber. The behavior of the mouse was registered for 1 minute.
After 6th day of experiment escape hole was transferred into 180°. Then followed 3 days of training and testing on day 10 of the experiment.
On the 6th and 10th days of the experiment, the time spent by the mouse searching for an escape hole was recorded, and the models of the search behavior of mice were also evaluated.
DNA and RNA extraction
DNA was isolated from the mice brain compartments and feces using the Proba-GS kit (DNA Technology, Russia) according to the protocol. RNA was isolated from the mice brain compartments using the ExtractRNA kit (Evrogen, Russia) according to the protocol. Qualitative analysis of isolated DNA and RNA preparations was performed by electrophoresis in 2% agarose gel in 1x TAE buffer.
Estimation of bacterial composition of the gut microbi-ome
To assess the bacterial composition of the gut microbiome, a quantitative PCR method was used according to the protocol [33]. The reaction was performed on a CFX96TM Real-Time System thermocycler (Bio-Rad, USA) with the reaction mixture containing 1 × qPCRmix-HS SYBR (Evrogen, Russia); 20 pM primer combination (forward and reverse) (Evrogen, Russia); 10 ng DNA template.
Measuring the amount of mtDNA damage
The amount of mtDNA damage was estimated by longrange PCR using the Encyclo-polymerase kit (Evrogen, Russia) according to the protocol we developed earlier [34].
Measuring of gene expression
cDNA was obtained using MMLV reverse transcriptase (Evrogen, Russia) on an Eppendorf Mastercycler personal thermocycler (Eppendorf, USA) according to the protocol. Quantitative PCR was performed on a CFX96TM RealTime System thermocycler (Bio-Rad, USA) using a qPCRmix-HS SYBR kit (Evrogen, Russia). 18s and Gapdh genes were used as references. List of primer sequences is presented in Table 2.
Table 2.
List of primer sequences.
| Forward primer sequence (5'-3') Reverse primer sequence (5'-3') | Reverse primer sequence (5'-3') | |
|---|---|---|
| 18s | CGGCTACCACATCCAAGGAA | GCTGGAATTACTGTGGCT |
| Gapdh | GGCTCCCTAGGCCCCTCCTG | TCCCAACTCGGCCCCCAACA |
| Gclc | GCAGCTTTGGGTCGCAAGTAG | TGGGTCTCTTCCCAGCTCAGT |
| Nfe2l2 | GTCTTCACTGCCCCTCATC | TCGGGAATGGAAAATAGCTCC |
| Pink1 | GAGCAGACTCCCAGTTCTCG | GTCCCACTCCACAAGGATGT |
| Trp53bp1 | TGCTCACCCTGGCTAAAGTT | AATGTCTCCTGGCTCAGAGG |
| Txnr2 | GATCCGGTGGCCTAGCTTG | TCGGGGAGAAGGTTCCACAT |
| Gpx1 | AGTCCACCGTGTATGCCTTCT | GAGACGCGACATTCTCAATGA |
| Brca1 | CGAGGAAATGGCAACTTGCCTAG | TCACTCTGCGAGCAGTCTTCAG |
| Ogg1 | TGAGCTGCGTCTGGACTTGGTT | CTCCGTCTGAGTCAGTGTCCAT |
| Sod2 | CAGACCTGCCTTACGACTATGG | CTCGGTGGCGTTGAGATTGTT |
| Tnf | TCTTCTCATTCCTGCTTGTGG | GGTCTGGGCCATAGAACTGA |
| p62 | GCCAGAGGAACAGATGGAGT | TCCGATTCTGGCATCTGTAG |
Statistical analysis
Statistical analysis was performed using Statistica 10 (StatSoft. Inc., Tulsa, OK, USA). mtDNA damage was quantified by using DNA Damage Calculator (Voronezh, Russia). For calculation of normalized expression and copy number of mtDNA, standard Bio-Rad CFX Manager software was used. Principal Component Analysis was performed in RStudio v.7.2.576 (Posit PBC, Boston, MA, USA) using ggfortify v.0.4.15 R package.
The results were expressed as means ± SEM. Analysis of the normality of the distribution using the Shapiro-Wilk test indicated that the distribution deviated from normality. Therefore, non-parametric methods of analysis were used. The paired Wilcoxon test was used to compare of cognitive parameters and bacterial composition of the gur microbiome the in the same groups of mice at different time points (4 months vs. 8 months of age). The MannWhitney U test was used to compare groups of mice of he same age with different genotypes (C57BL/6 vs. APP/PS1). The analysis of the frequency of different escape hole search strategies in mice in the Barnes maze was conducted using Chi-squared analyses.
Results
Cognitive parameters of mice
We assessed the cognitive performance of mice by the latency time to find the escape hole. 4 months-old C57BL/6 mice spent 28.9 ± 5.7 seconds searching for the escape hole, while APP/PS1 mice spent 48.7 ± 3.3 seconds (P < 0.01). Similar trends persisted at 8 months of age, with APP/PS1 mice spending 49% more time searching for the escape hole compared to non-transgenic mice, although the differences were not statistically significant (P = 0.08). The Wilcoxon pairwise test indicated a tendency for an age-dependent increase in search time for the platform in APP/PS1 mice, but the differences were not statistically significant (P = 0.06) (Figure 1A).
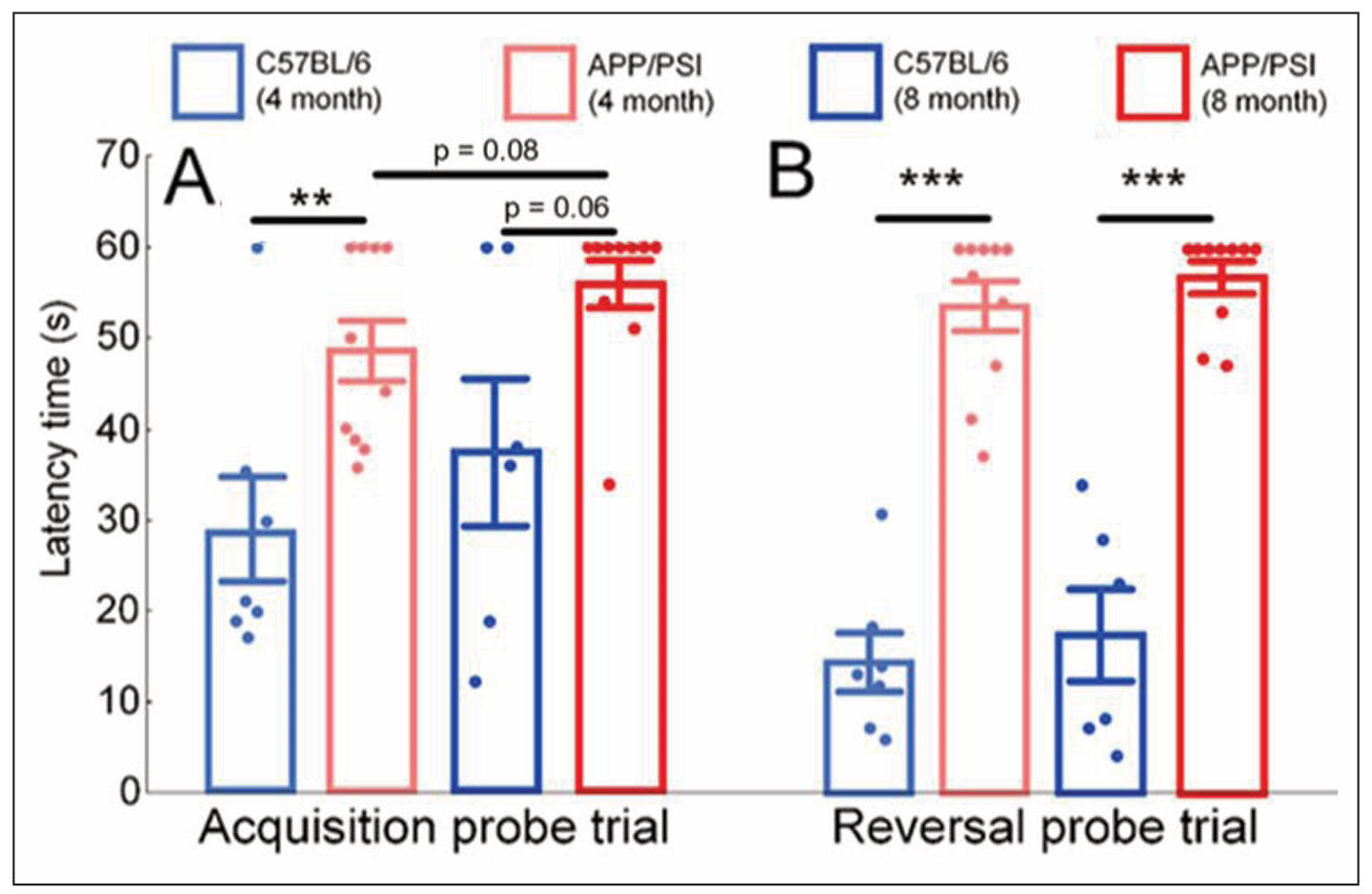
Figure 1. Results of the Barns maze test. Latency time to find the escape hole during acquisition (A) and reversal (B) probe trials for C57BL/6 and APP/PS1 mice strains. An increase in the time spent searching for the escape hole indicates a decline in cognitive performance of the mice. The results expressed as means ± SEM. The Mann-Whitney U test was used to compare groups of mice of the same age with different genotypes (C57BL/6 vs. APP/PS1), **P < 0.01, ***P < 0.001. The paired Wilcoxon test was used to compare the same groups of mice at different time points (4 months vs. 8 months of age).
After a 180° platform rotation, transgenic mice also demonstrated impaired cognitive abilities. At 4 months of age, control mice spent 14.4 ± 3.2 seconds searching for the platform, while transgenic mice took 53.6 ± 2.8 seconds (P < 0.001). Similar results were obtained at 8 months of age, with control mice spending 17.3 ± 5.1 seconds and APP/PS1 mice spending 56.8 ± 1.7 seconds searching for the platform (P < 0.001). Pairwise statistical analysis did not reveal age-related changes in platform search speed during the reversal probe trial (Figure 1B).
Search strategies significantly differ between APP/PS1 and C57BL/6 mice strains. The analysis of mouse search strategies allows us to identify four main models of mouse behavior. Direct strategy means moving directly to the target hole or to one-two adjacent holes before visiting the target escape hole (Figure 2A). Serial search strategy means subsequent visiting two or more holes before the escape hole. The search can be carried both clockwise and counterclockwise (Figure 2B). Mixed strategy means unorganized search, which includes crossing through the center of the maze (Figure 2C). Freezing is a behavioral feature which means lack of attempts to find the escape hole (Figure 2D).
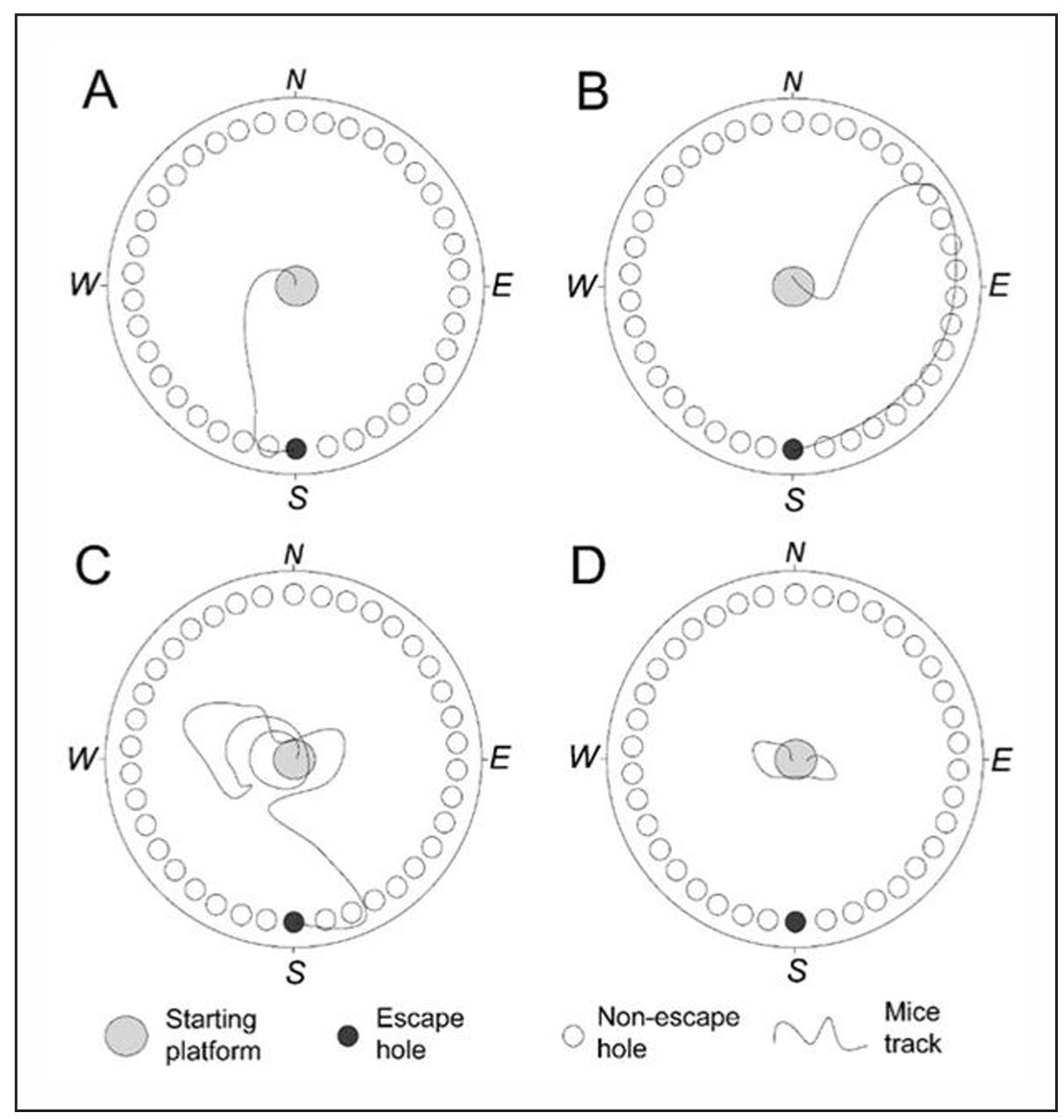
Figure 2. Representative examples of mouse tracks during different strategies of searching for the escape hole. (A) The direct strategy, where the mouse moves directly towards the target hole. (B) The serial search strategy, where the mouse sequentially investigates multiple holes one after the other. (C) The mixed strategy, where the mouse randomly moves around the maze. (D) Freezing, where the mouse remains near the starting platform and does not attempt to find the escape hole.
We found that 52% of 4-month-old C57BL/6 mice pre-fer serial search of the escape hole, whereas only 37% of APP/PS1 mice prefer this search strategy. In contrast, 38% of 4-month-old APP/PS1 mice prefer freezing behavior without an attempt to find the escape hole, while only 12% of 4-month-old C57BL/6 mice prefer this strategy (Figure 3A & B). Analysis of Pearson χ2 showed that differences in the search strategies between APP/PS1 and C57BL/6 mice were statistically significant (P = 0.0003), as well as between 8-month-old APP/PS1 and C57BL/6 mice (Pearson χ2, P = 0.00005). 8-month-old APP/PS1 mice prefer freezing behavior (49%), while only 15% of C57BL/6 mice did not make an attempt to find the escape hole. On the contrary, 41% of 8-month-old C57BL/6 mice moved directly to the target hole, while only 18% of APP/ PS1 mice used this strategy (Figure 3C & D).
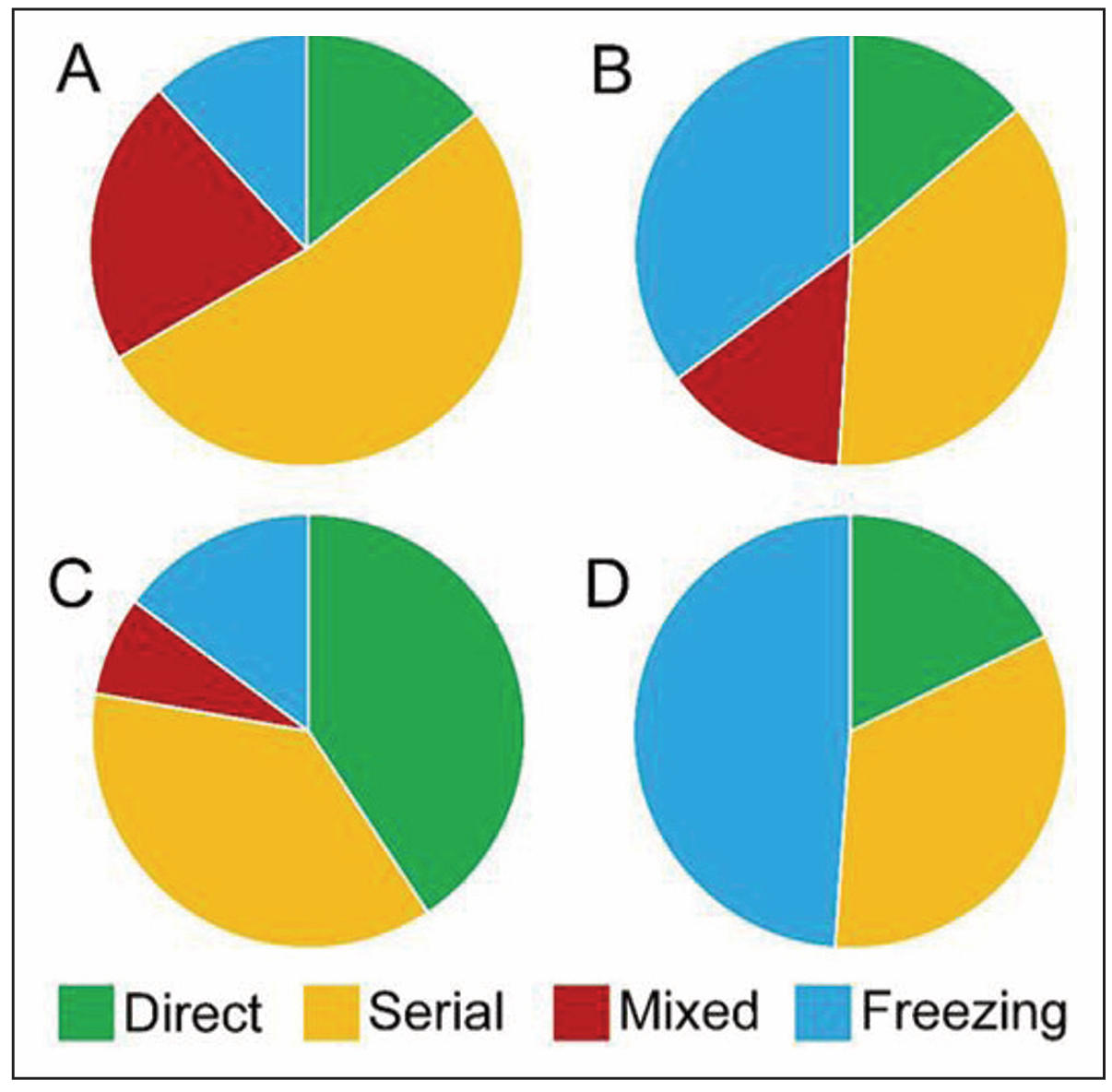
Figure 3. The frequency of occurrence of different escape hole search strategies. In 4-month-old C57BL/6 mice, the serial search strategy is predominant (A). Similarly, in 4-month-old APP/PS1 mice, the serial search strategy is prevalent, but a significant part of mice also exhibits freezing behavior (B). In 8-monthold C57BL/6 mice, both the direct and serial search strategies are prevalent (C). 8-month-old APP/PS1 mice exhibit a higher tendency towards freezing behavior without attempting to find the escape hole (D).
Bacterial composition of the gut microbiome
Using real-time PCR, we analyzed the ratio of the major bacterial phyla in the gut microbiome of C57BL/6 and APP/PS1 mice at 4 months and 8 months of age. The dominant phyla were Bacteroidetes and Firmicutes, which represented from 91% to 99% in the microbiome of each mouse. It is worth noting that no significant differences in the abundance of dominant phyla were observed between non-transgenic and transgenic mice. At 4 months of age, there were minimal differences in the gut microbiota composition between the two genotypes, except for significantly higher levels of Candidatus Saccharibacteria in APP/PS1 mice (P < 0.05). However, at 8 months of age, the transgenic mice had 3.5 times lower levels of compared to same-aged non-transgenic mice (P < 0.01). Additionally, at 8 months of age, the levels of Epsilonproteo-bacteria and Deltaand Gamma-proteobacteria in APP/ PS1 mice were higher than in C57BL/6 mice, although no statistically significant differences were observed (P = 0.058 for Deltaand Gamma-proteobacteria). Wilcoxon pairwise test revealed an age-dependent increase in Actinobacteria in both strains of mice, with a ninefold increase in C57BL/6 mice (P < 0.05) and a twofold increase in APP/PS1 mice (P < 0.01). Additionally, age-related decreases in Deferribacteres (fivefold decrease, P < 0.05) and age-related increases in Verrucomicrobia (twofold increase, P < 0.05) were observed in the transgenic mice.
Number of mtDNA damage in the brain compartments
We analyzed the number of damages in the six studied mtDNA fragments using long-range quantitative PCR. In the frontal cortex, we did not find any increase in the number of mtDNA damage in APP/PS1 mice compared with C57BL/6 mice. In contrast, C57BL/6 mice had twice more damage than APP/PS1 mice in the RNA non-coding region of the mtDNA D-loop (P < 0.01). A similar trend was observed in the hippocampus in the mtDNA fragment encoding ribosomal genes. The number of damages in the 12S/16S genes region was 73% higher in 8-monthold C57BL/6 mice compared to APP/PS1 mice (P < 0.01). In the midbrain, the number of damage in APP/PS1 mice compared with C57BL/6 mice twice increased in the mtDNA fragment encoding 16S and ND1 (P < 0.05) and in the D-loop region (P < 0.05) (Figure 4C). The greatest increase in the number of damage in APP/PS1 mice compared with C57BL/6 mice was observed in the cerebellum. The statistically significant increase in the number of damage was observed for the fragments encoding the 16S/ ND1 (+175%), ND1/ND2 (+112%), ND6/CytB (+146%), and the ND5 region (+239%) (all P < 0.05) (Figure 4).
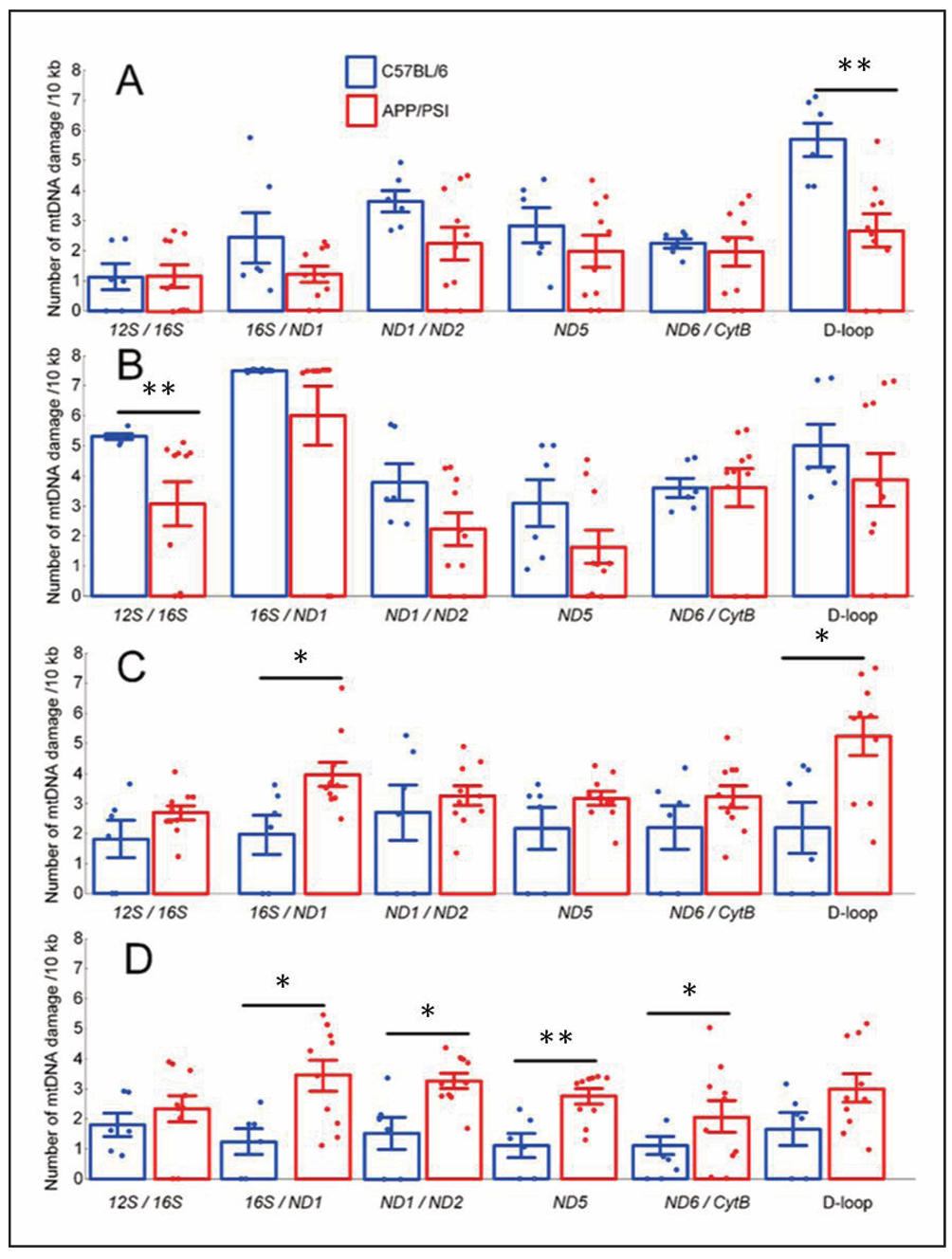
Figure 4. Number of mtDNA damage in the following brain regions: cortex (A); hippocampus (B); midbrain (C); cerebellum (D)in C57BL/6 and APP/PS1 mouse strains at 8 months of age. Results are expressed as mean ± SEM for each mtDNA fragment. *P < 0.05, **P < 0.01.
Gene expression in the brain compartments
We showed that the nuclear factor erythroid-derived 2-like 2 (Nfe2l2) gene expression was unchanged in the cerebral cortex, but tended to decrease in the hippocampus, midbrain, and cerebellum (Figure 5A). We did not find any statistically significant differences in the expression levels of breast cancer type 1 (Brca1) and 8-oxoguanine glycosylase (Ogg1) genes in any of the studied brain regions (Figure 5B & C). In APP/PS1 mice, p53-binding protein 1 (Trp53bp1) gene expression was threefold lower in the midbrain (P < 0.01) (Figure 5D).
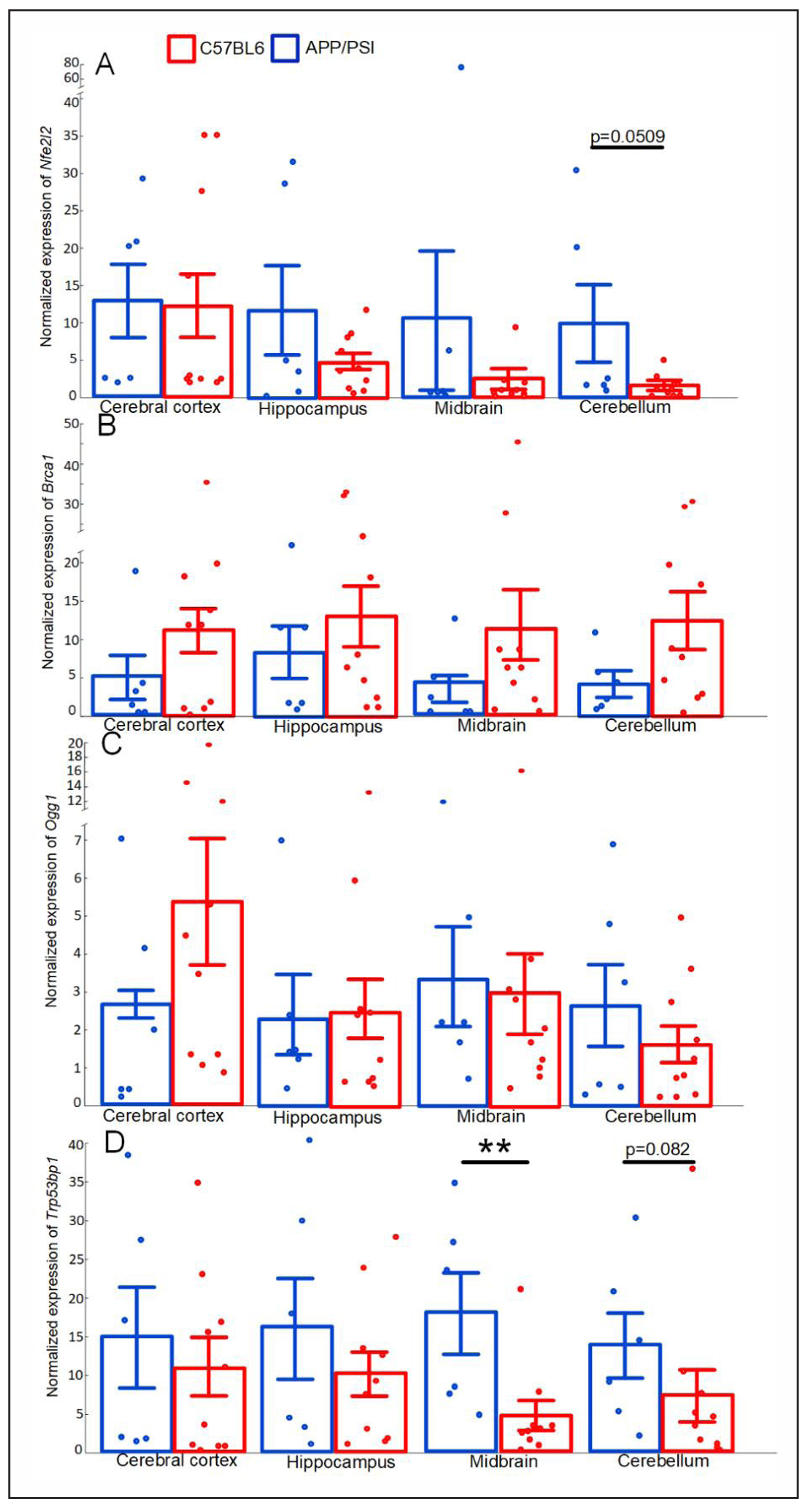
Figure 5. Normalized expression of Nfe2l2 (A), Brca1 (B), Ogg1 (C), Trp53bp1(D) in the cerebral cortex, hippocampus, midbrain, cerebellum of the C57BL/6 and APP/PS1 mice strains. The results expressed as means ± SEM. **P < 0.01 groups were compared using the Kruskal-Wallis test.
There were differences discovered in the expression of some antioxidant genes. The expression of glutathione peroxidase 1 (Gpx1) was approximately 5-fold lower in the cerebellum of transgenic mice compared to non-transgenic mice (P < 0.05). Similarly, the expression of the thioredoxin-2 (Txnr2) gene was decreased in the hippocampus and cerebellum in APP/PS1 mice (both P < 0.05) (Figure 6C). No statistically significant differences in the expression level of the superoxide dismutase 2 (Sod2) gene were found (Figure 6B).
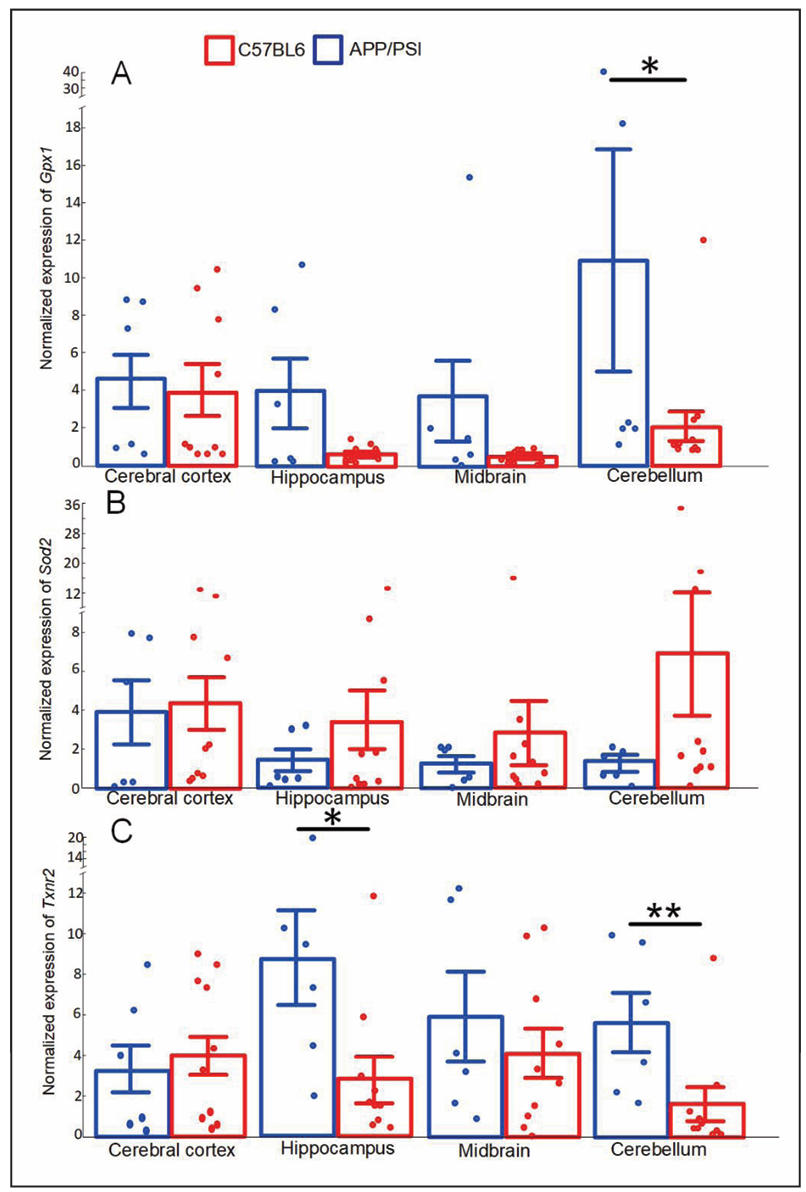
Figure 6. Normalized expression of Gpx1 (A), Sod2 (B), Txnr2 (C) in the cerebral cortex, hippocampus, midbrain, cerebellum of the C57BL/6 and APP/PS1 mice strains. The results expressed as means ± SEM. *P < 0.05, **P < 0.01 groups were compared using the Kruskal-Wallis test.
We did not find any differences in the expression of genes responsible for the regulation of mitophagy [sequestosome-1 (P62) and PTEN-induced kinase 1 (Pink1)] in the mice strains studied (Figure 7A & B). In APP/PS1 mice, the expression of the gene encoding the proinflammatory cytokine tumor necrosis factor α (TNF-α) was increased in the hippocampus (P < 0.05).
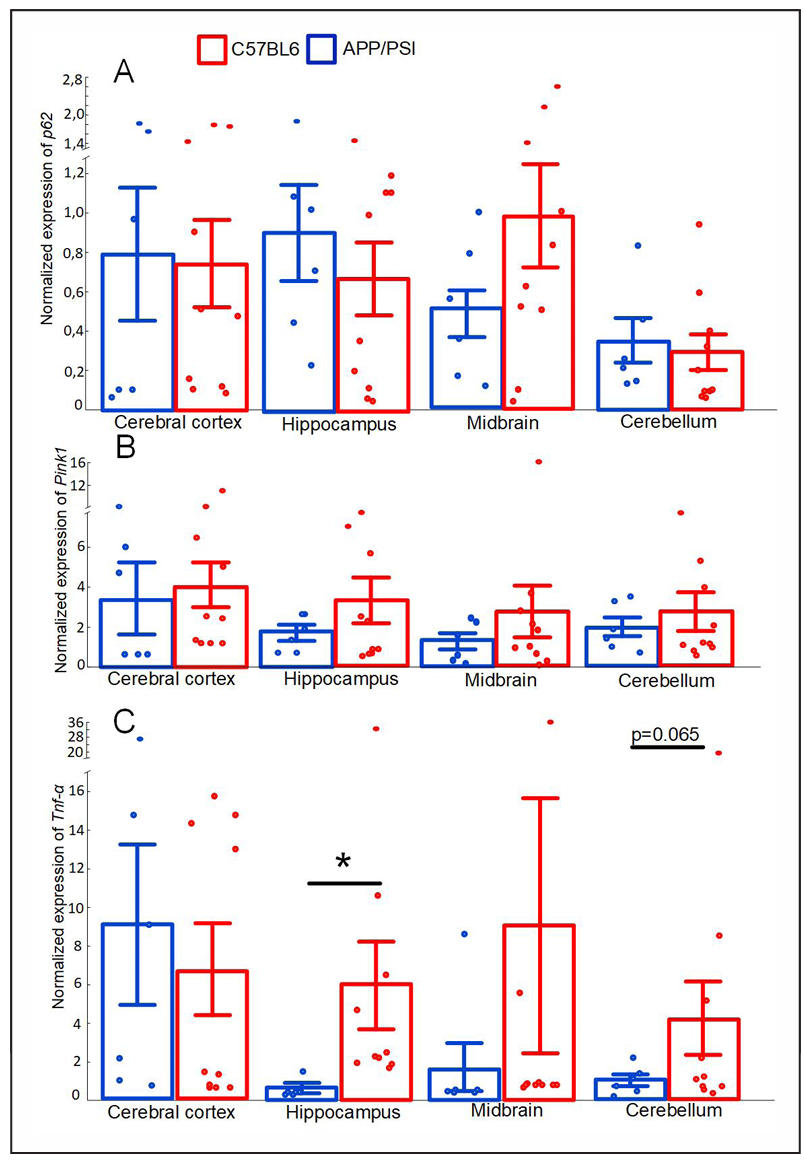
Figure 7. Normalized expression of P62 (A), Pink1 (B), Tnf (C) in the cerebral cortex, hippocampus, midbrain, cerebellum of the C57BL/6 and APP/PS1 mice strains. The results expressed as means ± SEM. *P < 0.05, **P < 0.01 groups were compared using the Kruskal-Wallis test.
Discussion
In recent years, AD research has emphasized the role of the brain, especially the neurodegenerative processes associated with oxidative stress, neuroinflammation and mitochondrial dysfunction. A growing body of evidence suggests that a key link in the pathogenesis of AD may be the dysregulation of antioxidant defense, in particular the Nrf2 pathway, which plays a critical role in maintaining cellular homeostasis. Nrf2/ARE is involved in many cellular processes that are associated with the maintenance of mtDNA integrity. We showed that in transgenic mice the decrease in the Nfe2l2 expression coincided with a significant decrease in the expression of Gpx1 and Txnr2 (Figure 6), which are involved in the glutathione and thioredoxin pathways of H2O2 utilization, respectively. Both glutathione and thioredoxin dependent systems dysfunctions are closely involved in increased oxidative stress in AD [35, 36].
Nrf2/ARE signaling pathway is not only involved in antioxidant protection of the cell, but is also necessary for maintaining the integrity of mtDNA by activating the elimination of damaged mitochondria through mitophagy [21] and triggering repair processes [37]. Nrf2 actually triggers mitophagy, but mitophagy can be triggered by damage to internal mitochondrial components, in particular mtDNA [38]. There were no differences detected in the expression levels of P62 and Pink1 genes, which are involved in the regulation of mitophagy (Figure 7А & B). We found a significantly reduced expression of the Trp53bp1 gene, which encodes the 53BP1 protein (Figure 5D) that plays a key role in the repair of double-stranded DNA breaks [39]. The presence of ARE sequences in the promoter region of the Trp53bp1 gene was previously shown in human colonic epithelial cells [18]. However, it is still unknown whether the mechanisms of double-strand break repair similar to those of nuclear DNA are typical for mtDNA. Base excision repair (BER) and mismatch repair (MMR) mechanisms for mtDNA are well described, but they only allow to repair damaged bases and mismatch bases only, but not double-stranded breaks. Some repair factors for double-strand breaks have been identified in mitochondria, but there are still no any convincing evidences that mtDNA is able to repair through the non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ), or homology-directed repair (HDR) pathways [40].
Nevertheless, a decrease in the Trp53bp1 expression in the midbrain and cerebellum of APP/PS1 mice coincides with an increase in mtDNA damage in these brain compartments (Figure 4C & D). Notably, no increase in mtDNA damage was detected in the cerebral cortex and hippocampus; instead, certain mtDNA fragments of APP/PS1 mice had less damage than mtDNA of C57BL/6 mice (Figure 4A&B) . The predisposition of midbrain dopaminergic neurons to accumulate mtDNA deletions has been shown previously [41], as confirmed by our data on the number of mtDNA damage in the midbrain (Figure 4C). The cerebellum has long been regarded as essential only for the coordination of voluntary motor activity and motor learning and has rarely been considered primarily in studies of AD pathogenesis [42]. However, postmortem analysis of the cerebellum of AD patients with the PS1-E280A mutation revealed greater Purkinje cell loss and more abnormal cerebellar mitochondria compared with controls [43]. This is supported by our study, which showed that 4 of the 6 mtDNA fragments of APP/PS1 mice studied had statistically significantly more damage than similar mtDNA fragments in C57BL/6 mice (Figure 4D).
Significant cerebellar damages may explain the peculiarities of mice’s task behavior in the Barnes test. We found that APP/PS1 at 8 months of age demonstrated the most pronounced freezing behavior (Figure 3D). It has been shown previously that suppression of cerebellar output may therefore facilitate freezing [44]. In addition, it is well known that cerebellar dysfunction is the cause of excessive daytime sleepiness [45]. We can suppose that it is cerebellar dysfunction that causes freezing behavior and decreased motivation, which explains why the average latency time to find the escape hole was significantly longer for APP/PS1 mice compared with C57BL/6 mice (Figure 1). It is worth noting that a statistically significant increase of latency time for APP/PS1 was observed only during reversal probe trials (Figure 1B), and partly during acquisition probe trials (Figure 1A). A similar observation of a deficit in the reversal phase of the Morris water maze spatial learning task was demonstrated in another study [45]. Thus, it is likely that APP/PS1 mice are significantly worse at switching attention to new tasks compared with C57BL/6 mice.
In recent years, interest in studying the relationship between the bacterial composition of the gut microbiome and cognitive impairment, including in AD, has increased considerably [28]. On the relationship between Actinobacteria levels and AD, contradictory data were previously obtained. On the one hand, APP/PS1 mice at 8 months of age have been shown to have decreased Actinobacteria levels compared with WT mice [46, 47]. In contrast, some studies have shown increased Actinobacteria levels in AD patients [48, 49]. Similar contradictory data have been reported for levels of Bifidobacterium, which belongs to the Actinobacteria type. In the American population of AD patients, Bifidobacterium levels are lower than those of healthy subjects [50], while, in contrast, in the Chinese population of people with AD, Bifidobacterium levels are higher than those of healthy people [51, 52]. Overall, studies agree that Proteobacteria levels are increased in AD, which has been repeatedly demonstrated in both AD patients [28, 41] and transgenic models [53]. We have demonstrated that Epsilon-, Delta-, and Gammaproteo-bacteria tend to increase in APP/PS1 mice compared with C57BL/6 at 8 months of age, but the differences are not statistically significant (Table 3). Increased Proteobacteria levels are one of the markers of dysbiosis development [54], which is one of the concomitant signs of the pathogenesis of neurodegenerative diseases, including AD [55]. However, we have demonstrated that levels are significantly lower in APP/PS1 mice compared with C57BL/6 mice. Earlier data on levels are also controversial. One study showed that transgenic mice were characterized by an increase in [53], whereas, in contrast, another study showed a decrease in levels [56].
Table 3.
List of primer sequences.
| Phylum | 4-month-old | M-U test | 8-month-old | M-U test | Wilcoxon pairwise test | |||
|---|---|---|---|---|---|---|---|---|
| C57BL/6 | APP/PS1 | P value | C57BL/6 | APP/PS1 | P value | C57BL/6 | APP/PS1 | |
| Bacteroidetes (%) | 86.70 ± 4.32 | 84.37 ± 1.28 | 0.130 | 89.40 ± 3.36 | 87.13 ± 3.01 | 0.551 | 0.345 | 0.333 |
| Firmicutes (%) | 11.37 ± 4.39 | 13.21 ± 1.08 | 0.088 | 7.55 ± 3.71 | 10.15 ± 2.56 | 0.481 | 0.173 | 0.333 |
| Actinobacteria (%) | 0.03 ± 0.01 | 0.10 ± 0.03 | 0.107 | 0.26 ± 0.10 | 0.48 ± 0.08 | 0.116 | 0.028 | 0.007 |
| Betaproteobacteria (%) | 0.55 ± 0.30 | 0.35 ± 0.08 | 0.591 | 0.66 ± 0.18 | 0.19 ± 0.03 | 0.008 | 0.249 | 0.114 |
| Epsilonproteobacteria (%) | 0.41 ± 0.16 | 0.22 ± 0.06 | 0.733 | 0.18 ± 0.06 | 0.29 ± 0.13 | 0.786 | 0.249 | 0.959 |
| Delta-and Gamma-proteobacteria (%) | 0.75 ± 0.38 | 1.30 ± 0.33 | 0.223 | 0.44 ± 0.27 | 1.47 ± 0.49 | 0.058 | 0.917 | 0.878 |
| Deferribacteres (%) | 0.15 ± 0.08 | 0.21 ± 0.06 | 0.306 | 0.03 ± 0.01 | 0.04 ± 0.01 | 0.871 | 0.249 | 0.017 |
| Candidatus Saccharibacteria (%) | 0.00 ± 0.00 | 0.21 ± 0.11 | 0.045 | 1.46 ± 0.33 | 0.25 ± 0.12 | 0.004 | 0.028 | 0.799 |
| Tenericutes (%) | 0.04 ± 0.02 | 0.03 ± 0.01 | 0.961 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.093 | 0.345 | 0.285 |
| Verrucomicrobia (ppm) | 0.11 ± 0.03 | 0.11 ± 0.01 | 0.591 | 0.27 ± 0.12 | 0.20 ± 0.05 | 0.871 | 0.116 | 0.037 |
Note: The results expressed as means ± SEM. The Mann-Whitney U test was used to compare groups of mice of the same age with different genotypes (C57BL/6 vs. APP/PS1). The paired Wilcoxon test was used to compare the same groups of mice at different time points (4 months vs. 8 months of age). P values indicated in the corresponding columns.
The gut-brain axis is a bidirectional system of interaction between the central nervous system and the gut, which is regulated at the neuronal, endocrine, metabolic, and im-mune levels. The intestinal mucosa makes initial contact with a large number of specific antigens of pathogenassociated molecular patterns (PAMPs) that are part of the bacterial cell wall. They are recognized by pattern recognition receptors (PRRs). Recognition of PAMPs by PRRs triggers the activation of several signaling pathways in the host immune cells, such as stimulation of various proinflammatory cytokines including TNF-α [57]. Numerous preclinical and clinical studies have shown excessive levels of TNF-α in the brain of AD patients and that TNFα-mediated neuroinflammation plays a key role in AD pathogenesis by enhancing Aβ production, decreasing Aβ clearance, increasing neuronal loss and cell death [58]. We revealed that the expression of the TNF-α encoding gene is increased in the hippocampus of APP/PS1 compared with C57BL/6 at 8 months of age (Figure 7C), which may indicate an increased level of inflammation in these brain compartments.
TNF-α exposure has a bimodal effect on the Keap1/Nrf2 system, and while an intense inflammatory activation suppresses expression of antioxidant proteins, a low level appears to be protective [59]. We found in the hippocampus, midbrain, and cerebellum of APP/PS1 mice a trend toward decreased expression of the Nrf2 encoding gene compared with C57BL/6 mice. The strongest decrease in the Nfe2l2 expression was observed in the cerebellum (P = 0.0577) (Figure 5A). Probably, inflammatory processes accompanying the pathogenesis of AD are one of the factors reducing the activity of the Nrf2/ARE signaling pathway. Cross-talk between Nrf2 and neuroinflammation in AD has been previously discussed [23].
Conclusions
It can be concluded that downregulation of the Nrf2/ARE pathway in APP/PS1 mice is most pronounced in the cerebellum and to a lesser extent in the midbrain. This is accompanied by decreased expression of a number of antioxidant genes, as well as genes responsible for DNA repair, which may be responsible for the increase in mtDNA damage. Cerebellar dysfunction mediated by the accumulation of mtDNA damage may explain the behavioral abnormalities we observed in Barnes’ maze. According to the data obtained, APP/PS1 mice may be significantly worse at switching attention to new tasks. Inflammatory processes, characterized by increased expression of proinflammatory cytokines and some changes in the composition of the gut microbiota, probably also contribute significantly to the development of cognitive and behavioral abnormalities in transgenic mice. The gut-brain axis is a bidirectional system of interaction between the central nervous system and the gut, regulated at different levels, which is important for further study of neurodegenerative diseases.
Declarations
Author contributions
Artem P. Gureev: Conceptualization, Methodology, Investigation, Data curation, Writing–original draft. Polina I. Babenkova: Investigation, Data curation, Writing–original draft. Veronika V. Nesterova: Investigation, Data curation. Arina D. Tsvetkova: Investigation, Data curation. Anna A. Eremina: Investigation. Vera A. Kryukova: Investigation. Irina S. Sadovnikova, Investigation.
Availability of data and materials
Data will be made available on request.
Financial support and sponsorship
The study was supported by the Ministry of Science and Higher Education of the Russian Federation within the framework of the State task for universities in the field of scientific activity (project No. FZGU-2023-0009).
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
1. Moskalev A, Guvatova Z, Lopes I, Beckett C, Kennedy B, De Magalhaes J, et al. Targeting aging mechanisms: pharmacological perspectives. Trends Endocrinol Metab, 2022, 33(4): 266-280. [Crossref]
2. Breijyeh Z, & Karaman R. Comprehensive review on Alzheimer’s disease: causes and treatment. Molecules, 2020, 25(24): 5789-5799. [Crossref]
3. Piaceri I, Nacmias B, & Sorbi S. Genetics of familial and sporadic Alzheimer’s disease. Front Biosci, 2013, 5(1): 167-177. [Crossref]
4. O’Brien R, & Wong P. Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci, 2011, 34: 185-204. [Crossref]
5. Zhang Y, Thompson R, Zhang H, & Xu H. APP processing in Alzheimer’s disease. Mol Brain, 2011, 4: 3-15. [Crossref]
6. Dewey C, Davies E, Xie H, & Wakshlag J. Canine cognitive dysfunction: pathophysiology, diagnosis, and treatment. Vet Clin North Am Small Anim Pract, 2019, 49(3): 477-499. [Crossref]
7. Thomsen B, Madsen C, Krohn K, Thygesen C, Schütt T, Metaxas A, et al. Mild microglial responses in the cortex and perivascular macrophage infiltration in subcortical white matter in dogs with age-related dementia modelling prodromal Alzheimer’s disease. J Alzheimers Dis, 2021, 82(2): 575-592. [Crossref]
8. Wirths O, & Zampar S. Neuron loss in Alzheimer’s disease: translation in transgenic mouse models. Int J Mol Sci, 2020, 21(21): 8144-8155. [Crossref]
9. Hall A, & Roberson E. Mouse models of Alzheimer’s disease. Brain Res Bull, 2012, 88(1): 3-12. [Crossref]
10. Dixit S, Fessel J, & Harrison F. Mitochondrial dysfunction in the APP/PSEN1 mouse model of Alzheimer’s disease and a novel protective role for ascorbate. Free Radical Biology and Medicine, 2017, 112: 515-523. [Crossref]
11. Emmerzaal T, Rodenburg R, Tanila H, Verweij V, Kiliaan A, & Kozicz T. Age-dependent decrease of mitochondrial complex II activity in a familial mouse model for Alzheimer’s disease. J Alzheimers Dis, 2018, 66(1): 75-82. [Crossref]
12. Phillips N, Simpkins J, & Roby R. Mitochondrial DNA deletions in Alzheimer’s brains: a review. Alzheimers Dement, 2014, 10(3): 393-400. [Crossref]
13. Jiang H, Niu F, Zheng Y, & Xu Y. CART mitigates oxidative stress and DNA damage in memory deficits of APP/PS1 mice via upregulating β-amyloid metabolism-associated enzymes. Mol Med Rep, 2021, 23(4): 11919. [Crossref]
14. Lovell M, Xiong S, Lyubartseva G, & Markesbery W. Organoselenium (Sel-Plex diet) decreases amyloid burden and RNA and DNA oxidative damage in APP/PS1 mice. Free Radic Biol Med, 2009, 46(11): 1527-1533. [Crossref]
15. Xu Y, Cheng L, Sun J, Li F, Liu X, Wei Y, et al. Hypermethylation of mitochondrial cytochrome b and cytochrome c oxidase II genes with decreased mitochondrial DNA copy numbers in the APP/PS1 transgenic mouse model of Alzheimer’s disease. Neurochem Res, 2021, 46(3): 564-572. [Crossref]
16. Xu Y, Xu L, Han M, Liu X, Li F, Zhou X, et al. Altered mitochondrial DNA methylation and mitochondrial DNA copy number in an APP/PS1 transgenic mouse model of Alzheimer disease. Biochem Biophys Res Commun, 2019, 520(1): 41-46. [Crossref]
17. Hahn A, & Zuryn S. Mitochondrial genome (mtDNA) mutations that generate reactive oxygen species. Antioxidants, 2019, 8(9): 392-405. [Crossref]
18. Fontana G, & Gahlon H. Mechanisms of replication and repair in mitochondrial DNA deletion formation. Nucleic Acids Res, 2020, 48(20): 11244-11258. [Crossref]
19. Choubey V, Zeb A, & Kaasik A. Molecular mechanisms and regulation of mammalian mitophagy. Cells, 2021, 11(1): 38-49. [Crossref]
20. Gureev A, Sadovnikova I, Starkov N, Starkov A, & Popov V. p62-Nrf2-p62 mitophagy regulatory loop as a target for preventive therapy of neurodegenerative diseases. Brain Sci, 2020, 10(11): 847-858. [Crossref]
21. Jayakumar S, Pal D, & Sandur S. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutat Res, 2015, 779: 33-45. [Crossref]
22. Ma Q. Role of Nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol, 2013, 53: 401-426. [Crossref]
23. Saha S, Buttari B, Profumo E, Tucci P, & Saso L. A perspective on Nrf2 signaling pathway for neuroinflammation: a potential therapeutic target in Alzheimer’s and Parkinson’s diseases. Front Cell Neurosci, 2021, 15: 787258. [Crossref]
24. Ahmed S, Luo L, Namani A, Wang X, & Tang X. Nrf2 signaling pathway: pivotal roles in inflammation. Biochim Biophys Acta Mol Basis Dis, 2017, 1863(2): 585-597. [Crossref]
25. Al Bander Z, Nitert M, Mousa A, & Naderpoor N. The gut microbiota and inflammation: an overview. Int J Environ Res Public Health, 2020, 17(20): 7618-7629. [Crossref]
26. Angelucci F, Cechova K, Amlerova J, & Hort J. Antibiotics, gut microbiota, and Alzheimer’s disease. J Neuroinflammation, 2019, 16(1): 108-112. [Crossref]
27. Hu X, Wang T, & Jin F. Alzheimer’s disease and gut microbiota. Sci China Life Sci, 2016, 59(10): 1006-1023. [Crossref]
28. Hung C, Chang C, Huang C, Nouchi R, & Cheng C. Gut microbiota in patients with Alzheimer’s disease spectrum: a systematic review and meta-analysis. Aging, 2022, 14(1): 477-496. [Crossref]
29. Abraham D, Feher J, Scuderi G, Szabo D, Dobolyi A, Cservenak M, et al. Exercise and probiotics attenuate the development of Alzheimer’s disease in transgenic mice: role of microbiome. Exp Gerontol, 2019, 115: 122-131. [Crossref]
30. Sun J, Xu J, Ling Y, Wang F, Gong T, Yang C, et al. Fecal microbiota transplantation alleviated Alzheimer’s diseaselike pathogenesis in APP/PS1 transgenic mice. Translational Psychiatry, 2019, 9(1): 189-199. [Crossref]
31. Tran T, Corsini S, Kellingray L, Hegarty C, Le Gall G, Narbad A, et al. APOE genotype influences the gut micro-biome structure and function in humans and mice: relevance for Alzheimer’s disease pathophysiology. Faseb j, 2019, 33(7): 8221-8231. [Crossref]
32. Gawel K, Gibula E, Marszalek-Grabska M, Filarowska J, & Kotlinska J. Assessment of spatial learning and memory in the Barnes maze task in rodents-methodological consideration. Naunyn Schmiedebergs Arch Pharmacol, 2019, 392(1): 1-18. [Crossref]
33. Yang Y, Chen M, Yang B, Huang X, Zhang X, He L, et al. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl Environ Microbiol, 2015, 81(19): 6749-6756. [Crossref]
34. Gureev A, Shaforostova E, Starkov A, & Popov V. Simplified qPCR method for detecting excessive mtDNA damage induced by exogenous factors. Toxicology, 2017, 382: 67-74. [Crossref]
35. Arodin L, Lamparter H, Karlsson H, Nennesmo I, Björnstedt M, Schröder J, et al. Alteration of thioredoxin and glutaredoxin in the progression of Alzheimer’s disease. J Alzheimers Dis, 2014, 39(4): 787-797. [Crossref]
36. Lasierra-Cirujeda J, Coronel P, Aza M, & Gimeno M. Betaamyloidolysis and glutathione in Alzheimer’s disease. J Blood Med, 2013, 4: 31-38. [Crossref]
37. Bártová E, Legartová S, Dundr M, & Suchánková J. A role of the 53BP1 protein in genome protection: structural and functional characteristics of 53BP1-dependent DNA repair. Aging, 2019, 11(8): 2488-2511. [Crossref]
38. Shu L, Hu C, Xu M, Yu J, He H, Lin J, et al. ATAD3B is a mitophagy receptor mediating clearance of oxidative stress-induced damaged mitochondrial DNA. Embo j, 2021, 40(8): e106283. [Crossref]
39. Kim S, Pandita R, Eskiocak U, Ly P, Kaisani A, Kumar R, et al. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proceedings of the National Academy of Sciences, 2012, 109(43): E2949-E2955. [Crossref]
40. Bender A, Schwarzkopf R, McMillan A, Krishnan K, Rieder G, Neumann M, et al. Dopaminergic midbrain neurons are the prime target for mitochondrial DNA deletions. J Neurol, 2008, 255(8): 1231-1235. [Crossref]
41. Jacobs H, Hopkins D, Mayrhofer H, Bruner E, van Leeuwen F, Raaijmakers W, et al. The cerebellum in Alzheimer’s disease: evaluating its role in cognitive decline. Brain, 2018, 141(1): 37-47. [Crossref]
42. Sepulveda-Falla D, Barrera-Ocampo A, Hagel C, Korwitz A, Vinueza-Veloz M, Zhou K, et al. Familial Alzheimer’s disease-associated presenilin-1 alters cerebellar activity and calcium homeostasis. J Clin Invest, 2014, 124(4): 1552-1567. [Crossref]
43. Vaaga C, Brown S, & Raman I. Cerebellar modulation of synaptic input to freezing-related neurons in the periaqueductal gray. Elife, 2020, 9: 54302. [Crossref]
44. Zhang L, Zhang J, Sun M, Chen H, Yan J, Luo F, et al. Neuronal activity in the cerebellum during the sleep-wakefulness transition in mice. Neurosci Bull, 2020, 36(8): 919-931. [Crossref]
45. Gallagher J, Minogue A, & Lynch M. Impaired performance of female APP/PS1 mice in the Morris water maze is coupled with increased Aβ accumulation and microglial activation. Neurodegener Dis, 2013, 11(1): 33-41. [Crossref]
46. Chu X, Hou Y, Meng Q, Croteau D, Wei Y, De S, et al. Nicotinamide adenine dinucleotide supplementation drives gut microbiota variation in Alzheimer’s mouse model. Front Aging Neurosci, 2022, 14: 993615. [Crossref]
47. Harach T, Marungruang N, Duthilleul N, Cheatham V, Mc Coy K, Frisoni G, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep, 2017, 7: 41802. [Crossref]
48. Ling Z, Zhu M, Yan X, Cheng Y, Shao L, Liu X, et al. Structural and functional dysbiosis of fecal microbiota in chinese patients with Alzheimer’s disease. Front Cell Dev Biol, 2020, 8: 634069. [Crossref]
49. Zhuang Z, Shen L, Li W, Fu X, Zeng F, Gui L, et al. Gut microbiota is altered in patients with Alzheimer’s disease. J Alzheimers Dis, 2018, 63(4): 1337-1346. [Crossref]
50. Vogt N, Kerby R, Dill-McFarland K, Harding S, Merluzzi A, Johnson S, et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep, 2017, 7(1): 13537. [Crossref]
51. Li B, He Y, Ma J, Huang P, Du J, Cao L, et al. Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement, 2019, 15(10): 1357-1366. [Crossref]
52. Zhou Y, Wang Y, Quan M, Zhao H, & Jia J. Gut microbiota changes and their correlation with cognitive and neuro-psychiatric symptoms in Alzheimer’s disease. J Alzheimers Dis, 2021, 81(2): 583-595. [Crossref]
53. Bäuerl C, Collado M, Diaz Cuevas A, Viña J, & Pérez Martínez G. Shifts in gut microbiota composition in an APP/PSS1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett Appl Microbiol, 2018, 66(6): 464-471. [Crossref]
54. Shin N, Whon T, & Bae J. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol, 2015, 33(9): 496-503. [Crossref]
55. Liu M, & Zhong P. Modulating the gut microbiota as a therapeutic intervention for Alzheimer’s disease. Indian J Microbiol, 2022, 62(4): 494-504. [Crossref]
56. Wang S, Jiang W, Ouyang T, Shen X, Wang F, Qu Y, et al. Jatrorrhizine balances the gut microbiota and reverses learning and memory deficits in APP/PS1 transgenic mice. Sci Rep, 2019, 9(1): 19575. [Crossref]
57. Takeda K, & Akira S. Toll-like receptors. Curr Protoc Immunol, 2015, 109: 14.12.11-14.12.10. [Crossref]
58. Chang R, Yee K, & Sumbria R. Tumor necrosis factor α Inhibition for Alzheimer’s Disease. J Cent Nerv Syst Dis, 2017, 9: 1179573517709278. [Crossref]
59. Shanmugam G, Narasimhan M, Sakthivel R, Kumar R, Davidson C, Palaniappan S, et al. A biphasic effect of TNF-α in regulation of the Keap1/Nrf2 pathway in cardiomyocytes. Redox Biol, 2016, 9: 77-89. [Crossref]