Open Access | Review
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Epigenetic mechanisms of the influence of aging on the development of breast cancer with the participation of retroelements
* Corresponding author: Rustam Nailevich Mustafin
Mailing address: Department of Medical Genetics and Fundamental Medicine,
Bashkir Medical University, 450008, Lenin street, 3, Ufa, Russia.
Email: ruji79@mail.ru
This article belongs to the Special Issue:Oxidative Stress and Mitochondrial Dysfunction in aging
Received: 29 May 2025 / Revised: 27 June 2025 / Accepted: 17 July 2025 / Published: 30 September 2025
DOI: 10.31491/APT.2025.09.180
Abstract
Breast cancer as a multifactorial disease is associated with aging and a variety of polymorphisms located mainly in intronic, regulatory and intergenic regions. Since retroelement genes are localized in these regions, it has been suggested that such polymorphisms lead to the development of breast cancer by altering the activity and functioning of retroelements. This assumption is also supported by the fact of pathological activation of retroelements during aging, which explains the increase in the incidence of breast cancer with age. This article describes the facts of activation and mechanisms of influence of LINE, SINE, LTR retroelements on breast cancer, which confirms the proposed assumption. In addition, the article describes the association of retroelement-derived oncogenic and tumor suppressor microRNAs involved in breast cancer carcinogenesis with aging, and reveals the mechanisms of interactions of retroelements with 86 microRNAs derived from them in these processes. The article describes promising ways of using the identified retroelement-derived microRNAs in targeted therapy of breast cancer using antisense oligonucleotides and the microRNAs themselves as tools. Using 17 of the retroelement-derived microRNAs described in the articles, the expression of which changes similarly in both breast cancer and aging, it is possible not only to target breast cancer therapy, but also to slow down aging.
Keywords
Long non-coding RNAs, carcinogenesis, microRNAs, breast cancer, retroelements, aging
Introduction
According to the latest data from the International Agency for Research on Cancer,
the age-standardized incidence rate of breast cancer (BC) is 46.2 per 100,000 population,
ranking first among all malignant neoplasms regardless of gender. The risk of developing
BC is associated with aging [1, 2]. In addition, BC treatments accelerate aging. Women
treated for BC suffer from higher rates of cognitive decline and physical
development [3, 4]. Therefore, it is promising to develop methods of targeted therapy aimed
at the mechanisms of breast cancer carcinogenesis without increasing aging, which meets the
principles of personalized medicine to ensure active longevity. About 10% of BC cases are
monogenic due to heterozygous germline mutations in tumor suppressor genes. 1/5 of such
monogenic forms of BC are caused by pathogenic mutations in the BRCA1 and BRCA2 genes,
the remaining 4/5 are caused by mutations in the ATM, BARD1, CHEK2, RAD51D, RAD51C, PALB2 genes.
However, most cases of BC are multifactorial diseases influenced by environmental factors and
hereditary predisposition under the influence of specific single nucleotide polymorphisms
(SNPs) [5]. According to the conducted meta-analysis of the results of GWAS (genome-wide
association study), 79 loci associated with BC were identified, among which 15 new SNPs
were studied, which are located in the regulatory, intronic or intergenic regions of
protein-coding genes. For example, rs72755295 is located in the intron of the EXO1
gene (encodes a protein for the repair of mismatched nucleotides), rs6507583 is in
the 18q12.3 region interacting with the SETB1 promoter [6]. In 2020, a meta-analysis
identified 32 new breast cancer susceptibility SNPs. Most of these SNPs are also located
outside genes or in their introns, with a putative effect on the regulation of downstream
genes or by an in trans mechanism as enhancers for genes such as TBX3, SOX4,
RNF115, POLR3C [7]. In 2023, GWAS have identified over 300 SNPs associated
with BC development [8]. A polygenic risk assessment including 313 SNVs has been developed
and to predict BC risk [9]. In 2025, based on the GWAS results 191 SNP-related genes significantly
associated with BC recurrence and metastasis [10]. In 2025, 332 independent association signals
for breast cancer were identified, including 131 previously known signals. Using integrated
functional genomics data analysis, 195 putative breast cancer susceptibility genes were identified,
enriched in the Wnt/beta-catenin, p53, TNF/NF-κB, PI3K/AKT pathways [11]. The location of SNPs in
gene introns, in intergenic and regulatory regions, is characteristic of most multifactorial
diseases [12] and is consistent with data indicating that these loci contain genes for retroelements
(REs), long non-coding RNAs (lncRNAs) and microRNAs [13]. Accordingly, BC-associated SNPs may
influence the activity of REs, lncRNAs, and microRNAs [14].
It should be noted that the genes of many non-coding RNAs (ncRNAs) originated from retroelements
(REs) in evolution or are formed by processing their transcripts [15]. An example of the
influence of SNP on epigenetic regulation of genes is the rs1972820 polymorphism in the 3’-UTR
(untranslated region) of the ERBB4 gene (encodes the tyrosine protein kinase receptor,
a member of the epidermal growth factor receptor family). Polymorphism rs1972820 alters binding
to the 3'-UTR of the gene with oncogenic microRNA miR-3144-3p (derived from LINE1 [15]), which
reduces the risk of breast cancer [16]. Another example is the SNP within the EXO motif: GGAG
and GCAG in the miR-1246 sequence (derived from ERVL-MalR [15]), which affects its intracellular
trafficking and stability [17]. This microRNA promotes metastasis and chemoresistance of breast
cancer cells by inhibiting the expression of the NFE2L3 gene (encodes a membrane-bound
glycoprotein of the nuclear envelope and endoplasmic reticulum) [18]. Since aging, which is
associated with breast cancer [1, 2], promotes progressive epigenetic derepression of REs [19],
this factor may be a trigger for the induction of retroelements altered under the influence of
breast cancer-associated SNPs for the development of BC carcinogenesis [6-11].
New directions in the study of epigenetic mechanisms of breast cancer development can become the
basis for more effective and safe methods of targeted therapy. Of greatest interest in this regard
is the study of the relationship between retroelements and epigenetic factors and aging in breast
cancer carcinogenesis. REs are mobile elements that move within the genome using a copy-and-paste
mechanism and occupy 46.67% of the human genome. The most common REs are autonomous non-LTR
(long terminal repeat) LINEs (long interspersed nuclear elements), occupying 21% of the genome,
while non-autonomous SINEs (short interspersed nuclear elements) account for 13%, and
LTR-containing REs account for 9% of the human genome [13]. Since many breast cancer-associated
polymorphisms located in loci of the human genome where retroelements are located can lead to
changes in REs activity, the aim of the study in this article is to describe experimental and
clinical data on the involvement of pathologically activated LINE, SINE and LTR retroelements.
Also for this purpose, the mechanisms of the influence of retroelements on the development of
breast cancer are described. In order to determine additional facts in favor of the role of
REs in breast cancer carcinogenesis, as well as to determine the significance of the study,
an analysis of the involvement of retroelement-derived microRNAs in the development of
breast cancer was carried out. The significance of identifying such microRNAs with oncogenic
and tumor suppressor properties lies in the possibility of using them as targets and tools,
respectively, for targeted therapy of breast cancer. Moreover, identifying specific
microRNAs derived from REs and involved in both aging processes and breast cancer
carcinogenesis will allow us to determine the mechanisms of the influence of aging on
the development of breast cancer and to determine the ways of targeting these mechanisms.
The impact of LINE1 on breast cancer development
Epigenetic changes leading to LINE1 activation, characterized by
hypomethylation of their loci, have been identified in breast cancer
patients [20]. Worse prognosis of breast cancer was also associated
with LINE1 hypomethylation [21]. In invasive breast cancer, LINE1
hypomethylation was correlated with negative ER status, and the
degree of LINE1 hypomethylation varied significantly across breast
cancer subtypes [22]. For LINE1 incapable of transposition (due to
truncation of the 5’-end) in breast cancer cells, their ability to bind
to transcription factors at their 3’-ends with a change in gene expression
was determined [23]. Using reverse transcriptase PCR, it was shown that
LINE1 expression suppresses breast cancer cell differentiation and promotes
lymph node metastasis [24]. A study of 7769 samples of various malignant
neoplasms showed an increase in the expression of retroelements in 3864 of
them, which correlated with the activation of 106 oncogenes with onco-exaptation
processes in half of the breast cancer samples [25]. Similar results were obtained
in another study with a study of 2954 samples of various tumors, when RE
insertions were detected in more than 50% of breast cancer tissues, mainly LINE1 [26].
Activated LINEs, in addition to activating oncogenes, can promote carcinogenesis by
inactivating tumor suppressors. This property has been noted for LINE1 in
relation to the WT1 [27], MCC [28], PTEN [29],
APC genes [30].
LINE activation in breast cancer stimulates carcinogenesis also by maintaining
telomere length in cell divisions with hTERT induction [31]. In addition, abnormally
expressed LINEs are sources of enzymes that retrotranspose non-autonomous retroelements
such as Alu, which are also involved in breast cancer carcinogenesis [32]. Activated
LINE1 in cancer promotes genomic instability and chromoanagenesis [33], while this
process is determined in more than 60% of cases of metastatic breast cancer [34]
and in more than half of HER2-positive breast cancer [35]. Thus, LINEs contribute
to breast cancer development through various pathways, including through
cross-regulation with their derived microRNAs (Figure 1).
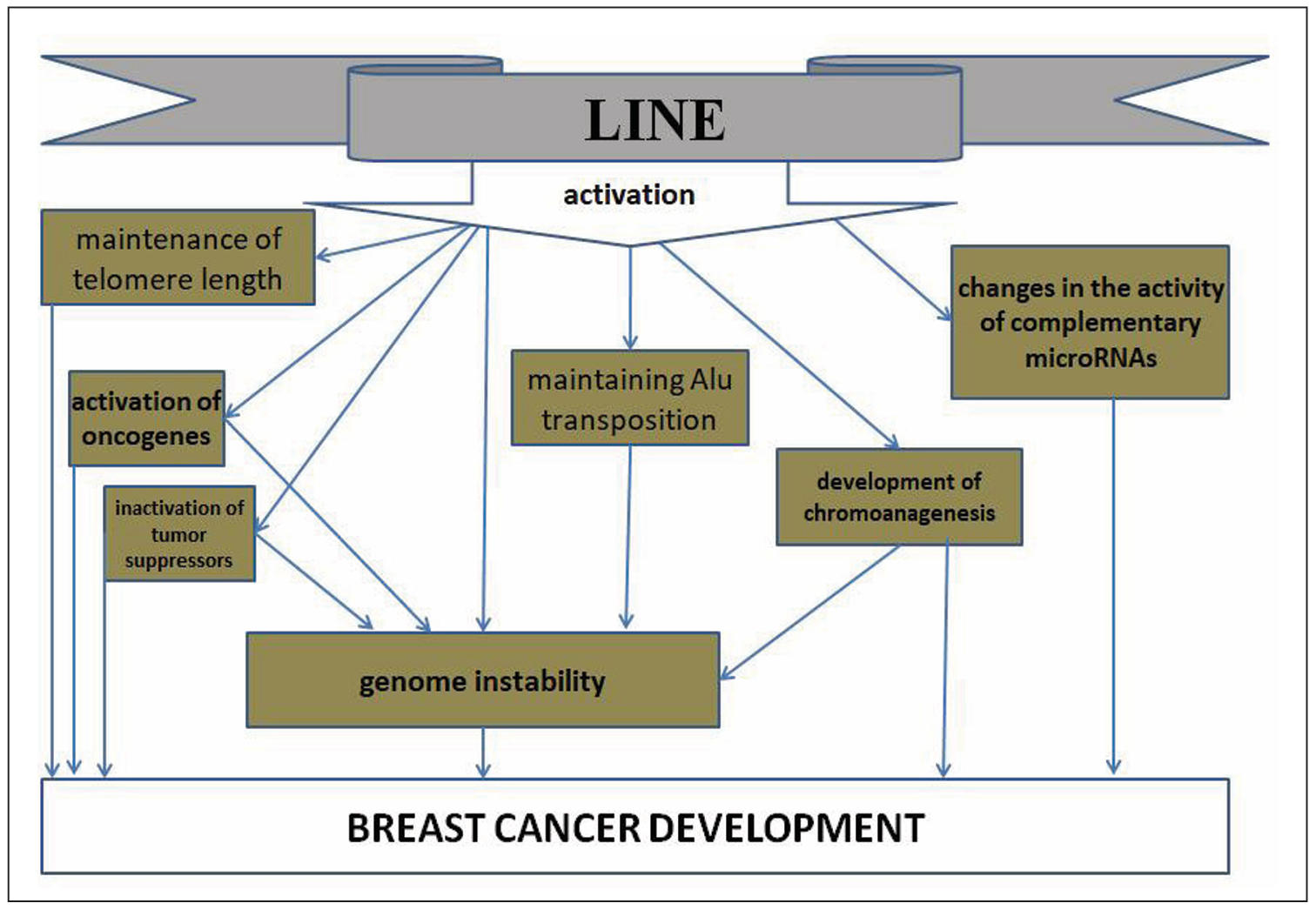
Figure 1. Pathways of LINE influence on breast cancer development.
The influence of SINE on the development of breast cancer
The most common SINEs in the human genome are Alu retroelements. Alu-PCR has
revealed multiple events of genomic instability mediated by Alu elements,
including deletions, insertions [36]. Under the influence of Alu, in breast
cancer carcinogenesis, inactivation of BRCA1, PIF, TSA,
ELG, RRM3 tumor suppressor genes often occurs [37]. In normal
cells, the CpG sites of Alu elements are methylated, which prevents their activation
and transposition [38]. Accordingly, hypomethylation causes activation of these REs.
In clinical studies, comparative analysis of Alu and LINE methylation in samples of
normal breast tissue, cells with atypia, ductal carcinoma in situ and invasive breast
cancer has determined a decrease in Alu methylation during the transition from
carcinoma in situ to invasive breast cancer, correlating with the negative status
of the estrogen receptor. In the HER2-enriched breast cancer subtype and with poor
patient survival, the lowest level of Alu methylation was detected [22]. In
metastatic breast cancer with early progression, hypomethylation of Alu was determined
in immune system cells. Moreover, an increase in the number of unmethylated loci was
associated with a worse prognosis [38]. A study of the epigenomic landscape of TNBC
tissues deficient in homologous recombination identified a decrease in Alu methylation
across the genome [39]. It should be noted that elevated levels of SINE are also
detected in the blood plasma of dogs with breast cancer, correlating with worse survival
compared to healthy animals. Also, increased expression of SINE B1 was detected in breast
cancer tissues at an early stage of tumor development in mice, with a marked increase
during cancer progression [40]. The obtained data indicate the universality of SINE as
a key player in breast cancer carcinogenesis in different species, including humans.
Promising biomarkers for breast cancer are circulating cell-free DNA, which is released
into the blood by tumor cells [41]. In clinical studies, a recognized effective method
for diagnosing and monitoring breast cancer is liquid biopsy, which can be used to detect
molecules that play a role in carcinogenesis in extracellular material [32]. The role of
actively expressed Alu in the development of breast cancer was also identified by
determining cell-free DNA ALU-247 and ALU-115, which levels were statistically
significantly higher in breast cancer patients compared to healthy controls.
These indicators have been proposed as diagnostic markers for the early detection
of breast cancer [42]. ALU-247 levels were significantly lower in the plasma of
breast cancer patients after surgery compared with preoperative data, and significantly
higher in patients with metastatic breast cancer [32]. Analysis of blood plasma samples
before and after a course of neoadjuvant chemotherapy in breast cancer patients with
determination of Alu-RNA levels by quantitative real-time PCR showed an increase in Alu
expression, more pronounced in premenopausal women and in hormone-positive breast cancer
[43, 44]. Analysis of circulating Alu also allows to assess their methylation status,
which was found to be higher in breast cancer patients compared to healthy controls [41].
Alu play an important role in the formation of circular RNAs (circRNAs) that are involved
in carcinogenesis, including breast cancer. It has also been determined that the homologous
splicing factor SLU7 regulates the formation of circCAPG circular RNA by binding to the
flanking Alu sequences of circCAPG transcripts (which are used to form circCAPG). This
circRNA promotes proliferation and metastasis of TNBC cells through the action of the
CAPG-171aa polypeptide, which is formed during translation of circCAPG on ribosomes
[45]. Inverted repeats of Alu (IRAlu) flank sequences of another circular RNA,
circRNA-CREIT. The RNA-binding protein DHX9 interacts with these IRAlu, suppressing
the expression of circRNA-CREIT, thus promoting the progression of doxorubicin-resistant
TNBC [46]. SINE expression products can stimulate the body's antitumor response as
inducers of the interferon response. This effect was determined using arginine
methyltransferase type 1 inhibitor, which promotes the interferon antitumor response
through antiviral defense pathways with a response to double-stranded RNA
(formed from inverted repeats of Alu elements) [47]. This may explain the described
phenomenon, when patients with low levels of circulating Alu and LINE were found to
have a lower risk of breast cancer recurrence [48].
In addition to identifying the fact of SINE activation in breast cancer,
the consequences of such events in the form of their integration into new loci of
the genome with disruption of the functions of tumor suppressor genes and the
transformation of proto-oncogenes into oncogenes were also determined. These
events can be drivers in breast cancer carcinogenesis. Thus, when examining breast
cancer samples in two independent studies, transpositions of retroelements, including
Alu, were identified in half of them [25, 26]. A genome-wide analysis of the landscape
of retroelement changes in breast cancer genomes conducted in 2024 using the ARTEMIS
(Analysis of RepeaT EleMents in dISease) approach showed changes in 73% of Alu and
97% of MIR sequences [49]. It can be assumed that such changes are recorded in the
results of the analysis of SNP associations with breast cancer [6-11], since these
SNPs are located mainly in introns and regulatory regions of genes where SINEs are
located (Figure 2).
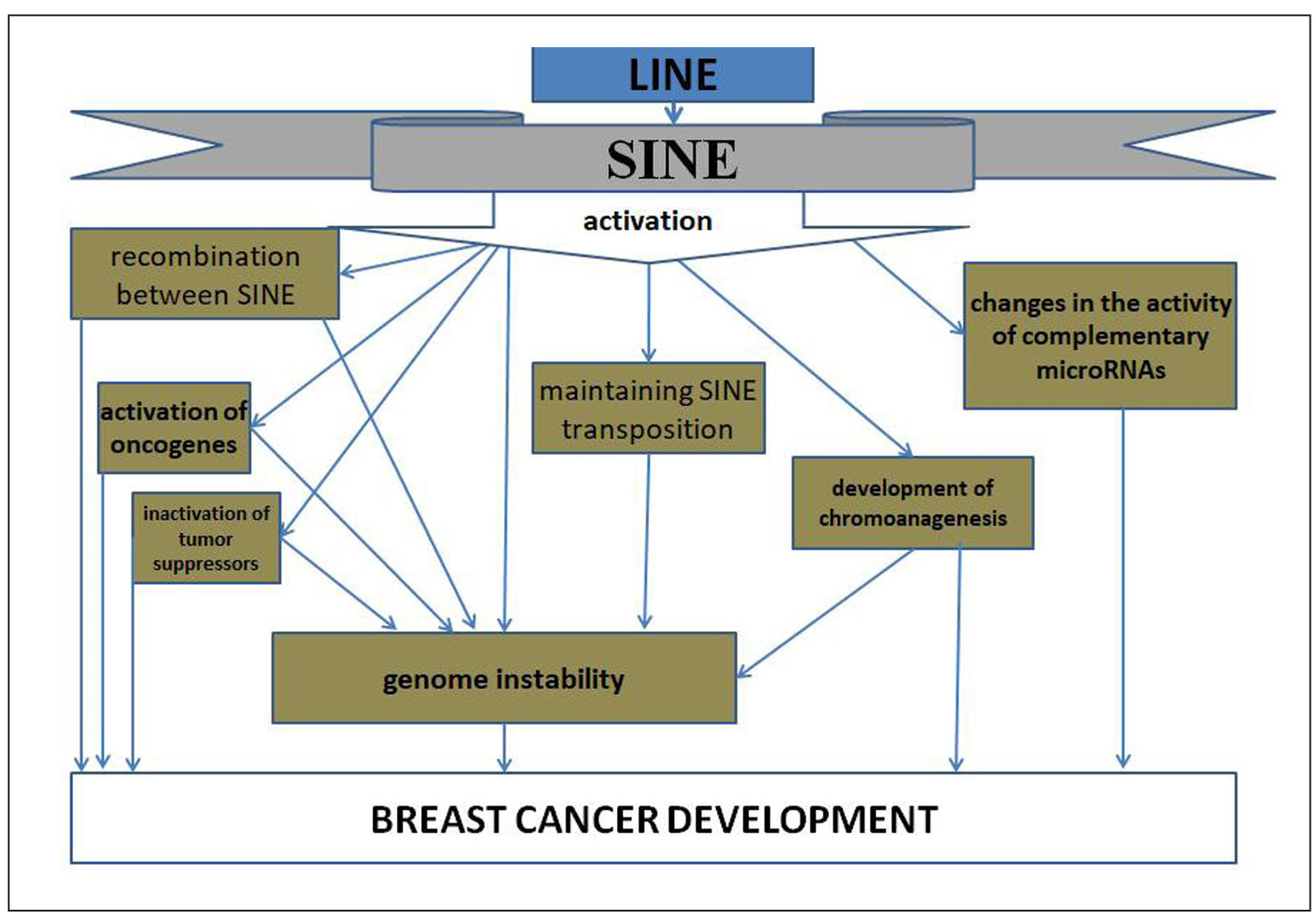
Figure 2. Mechanisms of influence of breast cancer-associated SNPs on the involvement of SINE in breast cancer carcinogenesis.
Effects of LTR retroelements on breast cancer development
Activation of LTR-REs is detected by direct identification of their expression in
breast cancer tissues, indirectly by identifying a decrease in methylation of their
loci in the genome, and also by analyzing cell-free DNA in the blood plasma of
patients. In breast cancer cells, HERV-K expression was found to be stimulated by
progesterone after exposure to estrogens [50], indicating a role for LTR-REs in
hormone-mediated breast cancer growth. Analysis of HERV expression in breast cancer
tissues using reverse transcriptase PCR showed the presence of HERV-K transcripts
(and the absence of HERV3, HERV-E4-1) in most breast cancer samples and cell lines,
indicating the influence of specific HERVs on the development of breast cancer.
Full-length proviruses and mature HERV envelope (env) mRNA were detected. No such
changes were found in normal breast tissue [51]. Further studies have shown that the
level of HERV-K expression in a breast cancer cell line increases 5-10-fold when
exposed to estradiol and progesterone [52], which may indicate a role for these
retroelements in the progression of hormone-dependent breast cancer. Experiments
on human T47D breast cancer cell lines revealed that the female sex hormones estradiol
and progesterone synergistically activate HERV-K via nuclear receptors. The progesterone
receptor isoform B binds to the progesterone response element (PRE) in the region of
the specific LTR5HS of HERV-K. In addition, there is another transcription factor
binding element OCT4, an octameric motif required by hormones to activate HERV-K
downstream of the LTR [53].
Suppression of HERV-K blocked the synthesis of Ras, p-RSK, p-ERK oncogenes that
affect breast cancer, and HERV-K overexpression restored the Ras/Raf/MEK/ERK
oncogenic pathways. HERV-K also activates CDK5, which suppresses the p53 tumor
suppressor by disrupting p53 phosphorylation. In turn, CDK5 is a mediator of
TGF-β1-induced epithelial-mesenchymal transition and tumor cell migration [54].
It should also be noted that 1509 LTRs in the human genome contain a nearly ideal
DNA binding site for the tumor suppressor protein p53, which exerts a regulatory
effect on them [55]. At the same time, mutations in the TP53 gene are
found in 40-60% of breast cancer samples [56], which serves as one of the additional
mechanisms of pathological activation of LTR-REs in breast cancer. Thus, breast
cancer is characterized by an increase in the expression of LTR-REs, which may
play an important role in the progression and clonal evolution of the tumor,
especially given the influence of female sex hormones on this process. Indeed,
a study of RE transcription in various types of breast cancer showed that the
expression of LTR sequences is characteristic of luminal-B HER2-positive breast
cancer [57]. Identification of cell-free DNA using whole-genome sequencing
allowed us to identify breast cancer-specific LTRs, as well as their insertions
near carcinogenesis driver genes. In breast cancer, changes were detected in 66%
of LTR-ERV1, 68% of LTR-ERVK, 66% of LTR-ERVL, 78% of LTR-ERVL-MaLR, and 67% of
LTR-Gypsy elements, indicating a global scale of genome transformations [49],
since LTR-REs play an important role in regulating the expression of many genes
during cell differentiation during development of the organism, since HERV
transcripts function as lncRNAs to control human embryonic stem cells [58].
LTR-REs activated in breast cancer exert their influence on carcinogenesis
in various ways. First of all, since they serve as regulators of cell differentiation
during ontogenesis [58], pathological expression of the evolutionarily youngest of
them, HERV-K, promotes dedifferentiation [59]. Since regulatory LTRs are globally
distributed in the human genome, serving as binding sites for various transcription
factors in the regulation of gene expression [60], their activation can cause
increased expression of downstream oncogenes. Indeed, it has been found that
activation of the oncogene IRF5 (interferon regulatory factor 5) is due to the
influence of the upstream LTR of this gene in Hodgkin's lymphoma [61]. Cases of
insertion of LTR-REs into the intron of the FABP7 proto-oncogene with
its transformation into the LTR2-FABP7 oncogene in B-cell lymphoma have
been described [62], insertion of an LTR-containing REs into the ERBB4
gene with the formation of the LTR-ERBB4 oncogene [63], and transposition
of LTR into the ALK gene with the formation of an alternative LTR-ALK
transcript [64]. In addition, tumor suppressor genes, including BRCA [65], are
characterized by the presence of hot spots for insertional carcinogenesis [66].
Therefore, activation of LTR-REs may cause decreased control of tumor suppressor genes.
Activated LTR-containing REs are potential sources of chromoanagenesis in
carcinogenesis [33]. Investigation of the causes of segmental duplications and
large genomic transformations has identified 30 different LTR-Res on 12 separate
chromosomes as the causes of these phenomena [67].
In vitro experiments on breast cancer cell lines (MCF-7, AU565, MDA-MB-231 and MB468)
showed that HERV-K activation causes a regulatory effect on neighboring genes.
Moreover, for different breast cancer cell lines, different changes in the expression
of HERV-K_1q23.3, HERV-K_22q11.23, HERV-K_9q34.11, HERV-K_14q32.33 and adjacent genes
were detected, including the CD48, IGLL1, SLAMF1,
SLAMF1 genes that positively correlate with immune infiltration of breast
cancer [68]. The HERV-K envelope protein Env has oncogenic properties in breast
cancer. Stimulation of its expression promotes epithelial-mesenchymal transition
of normal breast cells [69]. In the blood of patients with breast cancer, elevated
levels of the Env protein are detected not only of HERV-K, but also of endogenous
retroviruses HERV-H, HERV-P, HERV-R, which are reduced under the influence of
chemotherapy, to a greater extent than in patients receiving radiation therapy [70].
High levels of HERV-K expression are characteristic of the aggressive basal-like
subtype of breast cancer, suggesting the potential use of HERV as prognostic biomarkers
for breast cancer [71]. Antibodies to HERV-K are planned to be used for early
diagnosis of breast cancer [72]. Thus, there are various mechanisms of influence
of activated LTR-containing RE on the development of breast cancer, among which
the relationship with epigenetic factors, which include DNA methylation, histone
modifications and the influence of microRNA, is also important [66] (Figure 3).
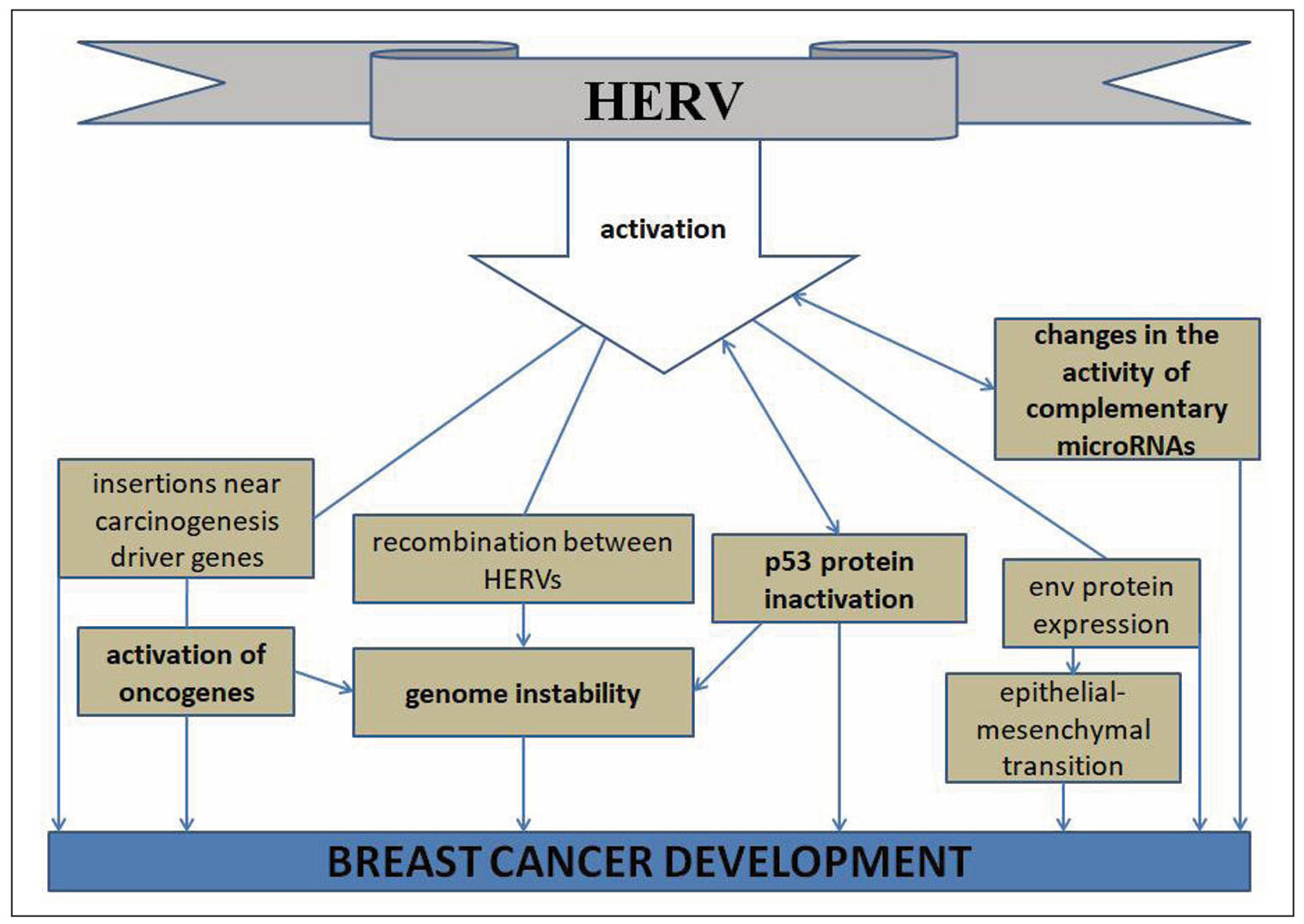
Figure 3. Mechanisms of carcinogenic action of HERV on breast cancer.
Association of retroelement-derived oncogenic microRNAs involved in breast cancer carcinogenesis with aging
The human genome contains 7,565 small noncoding RNA genes, most of which are microRNA genes [13]. According to a recent publication of the MDTE (miRNA derived from transposable elements) database [15], 474 microRNAs were found in humans that completely or partially overlap with sequences of mobile genetic elements. After excluding multicopy miRNAs, 405 unique mature miRNAs were identified, among which 352 completely overlapped with mobile genetic elements, accounting for 15% of the total number of molecules analyzed (2652 miRNAs). The richest sources of such microRNAs were DNA transposons (144 microRNAs), despite their insignificant share in the human genome. 116 microRNAs originated from LINE, 90 from SINE, and 50 microRNAs from LTR-RE [15]. It is possible that more microRNA genes derived from mobile genetic elements will be found in the future, since detailed genome analysis using specific oligonucleotides that recognize complementary mobile genetic elements (repeat sequences) has revealed that more than 2/3 of the human genome consists of such sequences [73]. The analysis of the microRNAs presented in the MDTE database with the data published in the scientific literature on the role of microRNAs in the development of breast cancer, as well as in the mechanisms of aging, allowed us to describe the microRNAs whose expression is increased (Table 1). Accordingly, microRNAs with increased expression exhibit oncogenic functions. Of the 43 microRNAs presented in the table, 17 microRNAs are characterized by changes in expression in specific tissues during human aging. The mechanism of the relationship between retroelements and oncogenic microRNAs in breast cancer may be due to the direct formation of microRNAs from transposable element genes [15]. Accordingly, increased expression of such microRNAs reflects the activation of such retroelements in breast cancer that inactivate tumor suppressor genes, promoting the inactivation of more tumor suppressor genes, thus enhancing carcinogenesis (Figure 4). In this regard, the use of such microRNAs as objects for targeted therapy of breast cancer (using antisense oligonucleotides) [74] in combination with reverse transcriptase inhibitors that suppress the transposition of activated retroelements is promising [75, 76].
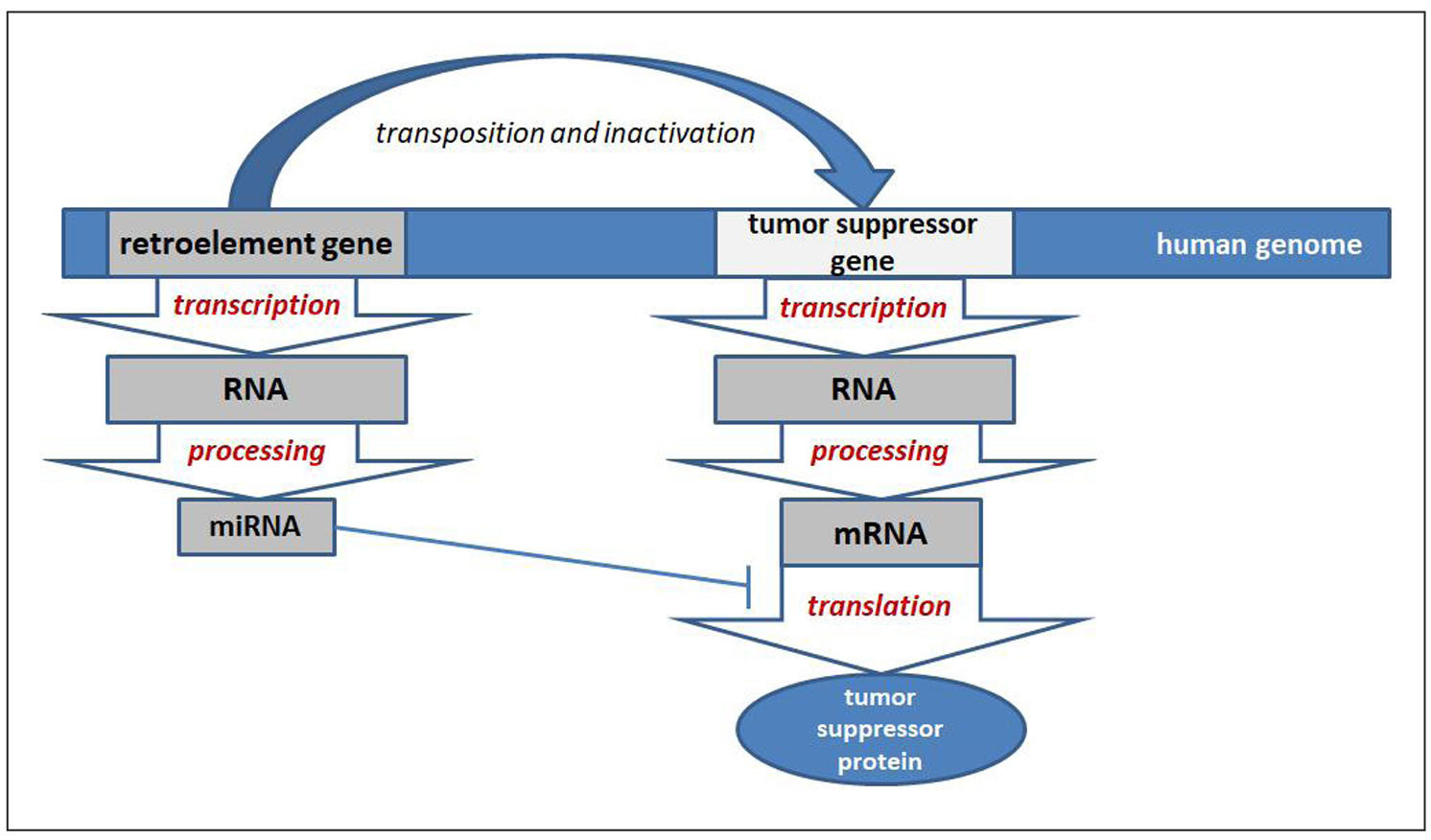
Figure 4. Scheme of the relationship between retroelements and oncogenic microRNAs in breast cancer carcinogenesis.
Table 1.
Retroelement-derived microRNAs which expression is increased in breast
cancer and altered with aging.
| No | RE-source of microRNA | microRNA/ tissue | miRNA target genes/interaction with other ncRNAs | changes in microRNA expression during aging (tissue) |
|---|---|---|---|---|
| 1 | SINE-MIR | miR-378d/ exosomes [77] | KLK4, NR2C2, NKX3-1, PHC3, ELAC1, ZNF124, RAB10, NCAPG, KCNIP2, VPS53, JADE3 (miRDB) | increased (colon tissue)[78] |
| 2 | LINE1 | miR-552-5p/ cancer [79] | WIF1/ lncRNA SLC16A1-AS1 [79] | increased (human epidermal cells) [80] |
| 3 | ERVL-MaLR | miR-558/ cancer [81] | HOXA1, CGREF1, ZNF217, EDC3, ULK1, ITGBL1 (miRDB) | increased (mesenchymal stem cells) [82] |
| 4 | LINE1 | miR-576-5p/ cancer [83] | MEIS2/ LINC-PINT [83] | increased (blood plasma) [84] |
| 5 | LINE2 | miR-1272-3p/ cancer [85] | MEF2A, DPYSL5, MTDH, USP3, WTAP, MAP3K20, ZCCHC10, STK35 (miRDB) | increased (human cerebral cortex) [86] |
| 6 | SINE-Alu | miR-3135b/ cancer [87] | LRRC27, FMNL3, TTC21B, CASTOR3, XPO7, PPM1A, DNM1L, RBP1 (miRDB) | increased (human hippocampus) [88] |
| 7 | LTR-ERV1 | miR-4428/ blood serum [89] | EPHB1, MECP2, KAT6A, ADAR, RELN, CCT8, KLF9, NCBP1 (miRDB) | increased (human salivary exosomes) [90] |
| 8 | SINE-MIR | miR-4480/ blood serum [89] | IL-17A [91] | increased (retina) [91] |
| 9 | SINE-MIR | miR-5100/ cancer [92] | AZIN1, MOXD1, SMARCA2, SPO11, MOSPD2, CSNK1G3, HTR2C (miRDB) | increased (brain microglia) [93] |
| 10 | LINE2 | miR-151a-5p/ blood [94] | UTY, FANCA, SEZ6L, AK2, RALGAPA1, APH1A, NDE1, RGS17 (miRDB) | decreased (serum) [95] |
| 11 | LINE2 | miR-325-3p/ cancer, breast cancer cell lines [96] | S100A2 [96], Trp53 [97] | decreased (human chondrocytes) [97] |
| 12 | SINE-tRNA-RTE | miR-342-5p/ breast cancer cell lines [98] | HER2 [98] | decreased (peripheral mononuclear cells) [99] |
| 13 | SINE-MIR | miR-378a-3p/ exosomes [77] | KLK4, NR2C2, NKX3-1, KIAA1522, PHC3, ELAC1, ZNF124, RAB10,NCAPG (miRDB) | decreased (thymus epithelial cells) [100] |
| 14 | SINE-MIR | miR-378a-5p/ cancer [101] | SUFU / lncRNA GAS5 [101] | decreased (human fibroblasts) [102] |
| 15 | LINE-CR1 | miR-582-5p/ cancer [103] | CMTM8 [103], FAM19A1 [104] | decreased (neural stem cells) [104] |
| 16 | ERVL-MaLR | miR-1246/ cancer [18] | NFE2L3 [18] | decreased (adipose tissue stem cells) [105] |
| 17 | ERVL | miR-1269a/ cancer [106] | CREBZF/ circPAPD4 [106] | decreased (ovaries) [107] |
| 18 | LINE2 | miR-95-3p/ cancer [108] | AKAP12/ circ_0001777 [109] | data not found |
| 19 | LINE2 | miR-151a-3p/ cancer [110] | FAM120AOS, AGO2, RPS6KA5, ME1, UPP2, YTHDF3, PITPNA (miRDB) | data not found |
| 20 | SINE-MIR | miR-378e/ cancer [111] | KLK4, NR2C2, NKX3-1, PHC3, ELAC1, ZNF124, RAB10, NCAPG, KCNIP2, VPS53, JADE3 (miRDB) | data not found |
| 21 | LTR-ERV1 | miR-548ay-3p/ breast cancer cell lines [112] | TMEM33, OGT, CCNA2, CDK17, SMC3, SNRPC, MIB1, CNOT7 (miRDB) | data not found |
| 22 | LINE2 | miR-616-3p/ cancer [113] | TIMP2 [113] | data not found |
| 23 | LINE2 | miR-887-3p/ cancer, extracellular vesicles [114] | BTBD7 [114] | data not found |
| 24 | LTR-ERV1 | miR-1202/ serum [115] | BICRAL, ZFHX4, CTNND1, TOX, RRN3, DMTF1, CRISPLD1 (miRDB) | data not found |
| 25 | Alu | miR-1268a/ cancer [116] | STXBP1, EEPD, EGR4, CMIP, FCAR, HPD, USB1, SLC8A2, TARP (miRDB) | data not found |
| 26 | ERVL | miR-1269b/ cancer [117] | URI1, DAAM1, AGAP1, CCL11, TRAF3 (miRDB) / lncRNA BRCAT54 [117] | data not found |
| 27 | Alu | miR-1285-3p/ cancer [118] | TP53, BTRC, SMURF1 [118] | data not found |
| 28 | Alu | miR-1304-3p/ cancer [119] | NR3C1, NFATC4, SCP2, ZMYM3, CNN3, PAGR1, PGM1 (miRDB) | data not found |
| 29 | ERVL-MaLR | miR-1587/ cancer [120] | IRF7 [120] | data not found |
| 30 | LINE2 | miR-1825/ serum [121] | CLCN3, NAA40, FUT9, DHX58, ABCA1, SNN, TUBG1 (miRDB) | data not found |
| 31 | ERVL-MaLR | miR-2909/ cancer [122] | MRE11, RAD50 [122] | data not found |
| 32 | LINE1 | miR-3118/ cancer [123] | ZMAT5, PPP1R7, RAB27A, MBNL1, KRAS (miRDB)/ lncRNA HAND2-AS1 [123] | data not found |
| 33 | LINE1 | MiR-3144/ cancer [16] | ERBB4 [16] | data not found |
| 34 | LTR-ERVL | miR-3200/ blood serum [124] | ZRANB2, INSYN2, FBN1, ANKRD10, AFTPH, REM2, PURA, CARF (miRDB) | data not found |
| 35 | SINE-MIR | miR-3617-3p/ cancer [125] | PPP2R2D, BMPR2, TET3, DAZ1, LPGAT1, NEXMIF (miRDB) | data not found |
| 36 | Alu | miR-3908/ cancer [126] | AdipoR1 [126] | data not found |
| 37 | SINE-MIR | miR-4425/ cancer [41] | PLEKHS1, PEG3, VWA5A, PDZD2, SNX19, MEX3C, RRM2B (miRDB)/ STXBP5-AS1 [41] | data not found |
| 38 | LINE2 | miR-4433b-5p/ extracellular vesicles [127] | CAMKK2, ABHD13, SOX6, DDX20, STK32A, NAT8L, CNTN1 (miRDB) | data not found |
| 39 | LTR-ERVK | miR-4448/ breast cancer cell lines [128] | PAN3, ARID2, BPTF, CHD2, SOX7, ZNFX1 (miRDB) | data not found |
| 40 | SINE-MIR | miR-5003-3p/ cancer [129] | CDH1 [129] | data not found |
| 41 | ERVL | miR-5694/ cancer [130] | AF9 [130] | data not found |
| 42 | LINE1 | miR-5698/ blood plasma [131] | MECP2, PPP1R9B, NFIX, WIZ, SLC6A17, CASTOR2, NR1D1, SRF (miRDB) | data not found |
| 43 | LINE1 | miR-8084/ blood plasma, cancer [132] | ING2 [132] | data not found |
Summarizing the general trends in Table 1,
among the REs-derived miRNAs that are upregulated in BC, 9 miRNAs (miR-378d,
miR-552-5p, miR-558, miR-576-5p, miR-1272-3p, miR-3135b, miR-4428, miR-4480,
miR-5100) are also upregulated in aging. Of these, 4 miRNAs are derived from
SINEs, 3 miRNAs from LINEs, and 3 from LTR-REs. That is, the uniform contribution
of different REs of the human genome in such relationships is determined. This
indicates that the activation of REs during aging can serve as a trigger for breast
cancer carcinogenesis not only by stimulating genomic instability, but also by forming
microRNAs with oncogenic properties from the transcripts of these activated REs.
8 microRNAs (miR-151a-5p, miR-325-3p, miR-342-5p, miR-378a-3p, miR-378a-5p, miR-582-5p,
miR-1246, miR-1269a) derived from REs (3 microRNAs derived from LINEs, 3 miRNAs
from SINEs, 2 miRNAs from LTR-REs) are characterized by increased expression in
breast cancer, but decreased expression in aging. This suggests that not only aging
is an inducer of altered REs expression leading to breast cancer development, but
also many other causes, including changes in epigenetic factors. This is also
evidenced by the increase in the levels of 26 other REs-derived microRNAs for
which changes in expression with aging have not been described in the scientific
literature.
Increased miR-95-3p expression in TNBC tissues is associated with worse patient
survival [108]. The target of miR-95-3p in these mechanisms is the mRNA of the
AKAP12 gene (the protein product of the gene enhances proliferation,
migration and invasion of breast cancer cells). The levels of miR-95-3p-interacting
circular RNA circ_0001777 are reduced in TNBC tissues. It is suggested that this
circular RNA be used in targeted therapy of TNBC [109]. In breast cancer, the most
pronounced increase in expression is determined for miR-151a-3p [110]. Among circulating
microRNAs in patients with breast cancer, elevated levels of miR-151a-5p were identified,
which decreased after surgical treatment of breast cancer [94]. Significantly Increased
Levels of miR-325-3p Targeting S100A2 mRNA are detected in breast cancer tissues and
cell lines [96]. MiR-342-5p is a potential regulator of HER2 (human epidermal growth
factor 2) in breast cancer [98]. Exosomal miR-378a-3p and miR-378d contribute to the
development of breast cancer and chemoresistance by activating EZH2/STAT3 signaling
[77]. MiR-378a-5p is the target of GAS5 lncRNA, which promotes breast cancer apoptosis.
The target of miR-378a-5p is the mRNA of the SUFU gene [101]. MiR-378e expression
is associated with worse survival in breast cancer patients treated with palbociclib.
It is suggested that miR-378e contributes to resistance to this drug in breast cancer
cells [111].
Rhythmic changes in miR-548ay-3p expression in breast cancer cell lines were
identified [112]. Oncogenic miR-552-5p targeting WIF1 (Wnt inhibitory factor-1)
is a target of lncRNA SLC16A1-AS1, the overexpression of which suppresses breast
cancer growth and metastasis [79]. Increased expression of miR-558 was detected in
TNBC tissues [81]. MiR-576-5p, which is a target of the tumor suppressor LINC-PINT,
inhibits MEIS2 (Meis homeobox 2), which suppresses PPP3CC (protein phosphatase 3
catalytic subunit gamma) by inactivating nuclear factor κB (NF-κB) [83]. MiR-582-5p
promotes TNBC metastasis and invasion by inhibiting CMTM8 (chemokine-like factor)
[103]. MiR-616-3p promotes breast cancer metastasis by inhibiting the TIMP2
gene (encodes a family of inhibitors of matrix metalloproteinases) with regulation
of MMP (matrix metalloproteinase) signaling [113]. Increased levels of miR-887-3p
targeting BTBD7 were detected in breast cancer cells and extracellular
vesicles. At the same time, increased expression of BTBD7 abolished chemo-resistance
of breast cancer cells [114].
MiR-1202 levels are elevated in the serum of breast cancer patients [115].
Tamoxifen-resistant LCC9 breast cancer cells express elevated levels of HNRNPA2/B1
(heterogeneous nuclear ribonucleoprotein A2/B1), which is a reader of the
N(6)-methyladenosine tag in primary microRNAs (pri-miRNAs), promoting the processing
of DROSHA into microRNA precursors (pre-miRNAs). In these mechanisms, HNRNPA2/B2
upregulates miR-1268a expression [116]. MiR-1269a, targeting CREBZF (negative-regulatory
transcription factor) mRNA, is a target of tumor suppressor circPAPD4 in breast
cancer [106]. Oncogenic miR-1269b is a target of lncRNA BRCAT54 (breast cancer-associated
transcript 54), which suppresses breast cancer development [117]. A study of TNBC tissues
showed increased expression of miR-1272-3p, involved in the regulation of thyroid hormone
signaling pathways. Elevated miR-1272-3p levels are associated with TNBC recurrence [85].
MiR-1285-3p, a target of circ_0000376, promotes breast cancer carcinogenesis via inhibition
of TP53, BTRC, SMURF1 genes [118].
It was found that exosomal miR-1304-3p promotes the development of breast cancer by
activating breast cancer-associated adipocytes [119]. MiR-1587, an inhibitor of
IRF7 gene mRNA (encoding interferon regulatory factor 7), promotes breast cancer
development by regulating M2 macrophage activity [120]. Serum miR-1825 levels in breast
cancer patients before surgery are significantly higher compared to healthy controls and
patients after mastectomy [121]. MiR-2909 is involved in breast cancer development by
inhibiting the RAD50 (involved in the repair of double-strand DNA breaks) and
MRE11 (required for homologous recombination and DNA repair) genes [122].
MiR-3118, an inhibitor of PHLPP2 (serine-threonine phosphatase), is a target
of lncRNA HAND2-AS1, which suppresses breast cancer cell proliferation and migration
[123]. Apocrine cell morphology of TNBC is characterized by increased expression of
miR-3135b, which is involved in the regulation of Wnt signaling, MAPK, endocytosis,
and ErbB signaling genes [87]. MiR-3200 levels elevated in breast cancer were found
to be reduced by neoadjuvant chemotherapy [124].
Increased expression of miR-3617-3p was detected in breast cancer cells resistant to
antitumor therapy [125]. MiR-3908 is a key mediator of breast cancer cell migration,
enhancing their clonogenic process. The direct target of miR-3908 is AdipoR1, through
the suppression of which SIRT-1, p-AMPK and AMPK are inhibited [126]. Oncogenic miR-4425
is a target of lncRNA STXBP5-AS1 in breast cancer carcinogenesis [41]. MiR-4428 expression
is associated with breast cancer metastases in the brain [89]. The levels of miR-4433b-5p
in extracellular vesicles of breast cancer samples are higher than in the normal control
[127]. The association of increased miR-4448 expression with the development of breast
cancer chemoresistance has been determined [128]. The association of elevated levels of
miR-4480 with breast cancer metastases was revealed [89]. MiR-5003-3p has been identified
to promote epithelial-mesenchymal breast cancer transition by directly inhibiting the
CDH1 (E-cadherin) gene [129]. A significant increase in miR-5100 was detected
in breast cancer tissues compared to normal samples [92]. MiR-5694, targeting the mRNA
of the AF9 protein (metastasis suppressor), promotes the progression and metastasis of
basal-like breast cancer [130]. Increased expression of miR-5698 in patients' serum is
associated with worse survival in breast cancer [131]. In the blood plasma and tumor
tissues of breast cancer patients, a significant increase in miR-8084 levels was determined,
which promotes the proliferation of breast cancer cells by activating AKT and ERK1/2,
and also inhibits apoptosis by suppressing p53-BAX-related signaling pathways. The
target of miR-8084 is also the tumor suppressor gene ING2 [132].
It should be noted that among the listed 43 oncogenic microRNAs derived from REs and
involved in breast cancer carcinogenesis, for 17 microRNAs (Table 1),
a decrease in expression was noted for 7 microRNAs (miR-151a-3p, miR-325-3p,
miR-342-5p, miR-378a-3p, miR-378a-5p, miR-582-5p, miR-1246, miR-1269a),
which is logical, since with age the proliferative potential of cells
decreases with a decrease in the activity of oncogenic molecules. Accordingly,
data for BC with aging are characterized by abnormal activation unrelated to
aging mechanisms, but associated with the functioning of the REs from which
they originated. In particular, miR-151a-5p levels are reduced in serum during
physiological aging [95], while in breast cancer the levels of the same microRNA
in the blood are increased [94]. Mechanical overload downregulates miR-325-3p
expression in chondrocytes, promoting their aging and worsening lumbar facet
joint degeneration via Trp53 inhibition [97]. SIRT6 (Sirtuin 6) is a deacetylase
that affects the structure of chromatin and slows down the aging of the human body.
A study of young, middle-aged and old people did not show age-related differences
in methylation of SIRT6 CpG islands. However, in long-lived people, a significant
decrease in the expression of miR-342-5p in peripheral mononuclear cells was determined
[99]. Experiments have shown that decreased expression of miR-378a-3p promotes thymus
involution and aging [100]. MiR-378a-5p is a negative regulator of aging [102]. With
aging, the ability of neural stem cells to proliferate decreases due to epigenetic
mechanisms, including disruption of microRNA expression. It has been shown that
miR-582-5p in these mechanisms helps maintain the proliferation of neural stem
cells, preventing brain aging by inhibiting the secretory protein FAM19A1 [104].
Exosomes overexpressing miR-1246 from adipose-derived stem cells exhibit
photoprotective effects against UV-induced photoaging. miR-1246 targets
GSK3β gene mRNA. Accordingly, decreased expression of this microRNA
contributes to aging [105]. MiR-1269a expression was found to be higher in
the ovaries of young women compared to older women [107]. It can be speculated
that during aging, increased expression of REs contributes to the decrease in the
levels of these microRNAs due to complementary binding of their sequences as
"sponges". However, oncogenes can induce senescence in response to hyperactive
oncogenic signals, since this develops protection against tumor development.
Experiments on human diploid fibroblasts have shown that activated miR-378a-5p
promotes the preservation of their proliferative capacity even in the presence
of the Braf oncogene, delaying oncogene-induced senescence. This microRNA reduces
the expression of senescence-associated β-galactosidase and p16INK4A [102]. This
example demonstrates the risks of using new methods to combat aging, since the side
effects of using specific microRNAs that can slow down aging include mechanisms to
stimulate the development of tumors, such as breast cancer. This demonstrates the
subtle mechanisms of epigenetic regulation of carcinogenesis and aging, which are
only partially related. And when using microRNAs as objects or tools for rejuvenation
of the human body, it is first necessary to assess the risks of cancer development.
At the same time, with aging, an increase in the expression of 9
microRNAs (miR-378d, miR-552-5p, miR-558, miR-576-5p, miR-1272-3p, miR-3135b, miR-4428,
miR-4480, miR-5100) was determined, the expression of which is increased in breast
cancer, which indicates that epigenetic mechanisms of aging can contribute to
carcinogenesis, in particular the development of breast cancer. A possible reason
is the hyperactivation of REs, since microRNAs can be transcribed not only from
their own genes, but also directly from REs transcripts [15]. For example, in colon
tissues, increased expression of miR-378d has been identified with aging [78]. In
skin aging, elevated calcium concentration in the basal layer reduces cell proliferation
with increased expression of miR-552-5p. Under the influence of lncRNA SPRR2C, which
binds complementarily to this microRNA, this aging effect is suppressed [80]. Increased
expression of miR-558 is detected in aging mesenchymal stem cells [82]. Elevated plasma
miR-576-5p levels detected in frail elderly compared to young adults [84]. Increased
mR-1272-3p levels in the cerebral cortex have been identified during physiological aging
in humans [86]. About 80% of differentially expressed genes in the hippocampus are
regulated by miR-192-5p and miR-3135b, which are elevated in older adults. However,
miR-3135b expression in the hippocampus was reduced in centenarians, suggesting that
miR-3135b contributes to accelerated aging [88]. In saliva exosomes of elderly people,
significantly more pronounced (2-fold) expression of miR-4428 was determined compared
to young people [90]. An example of the influence of SNP on the characteristics of
human aging is rs7747909 in the 3'-UTR of the IL-17A gene, which affects the
binding of miR-4480 to it, predisposing to the development of retinal degeneration
with aging [91]. Increased expression of miR-5100-3p detected in brain microglia during
aging [93].
Association of retroelement-derived tumor suppressor microRNAs involved in breast cancer carcinogenesis with aging
In the development of breast cancer, a decrease in the expression of a number of microRNAs has been noted, which have tumor suppressor properties, since they inhibit oncogene mRNA. Accordingly, such microRNAs can serve as tools for targeted therapy of breast cancer. Analysis of the MDTE database [15] revealed 43 REs-derived miRNAs that are downregulated in breast cancer. This indicates unique epigenetic mechanisms of carcinogenesis, when REs activation promotes breast cancer development, while REs-derived microRNAs have tumor suppressor properties. It can be assumed that one of the mechanisms of antitumor action of such microRNAs, in addition to inhibition of oncogenes, is competitive binding to REs transcripts, leveling their initiating effect on carcinogenesis (due to promoting genomic instability, inactivation of tumor suppressors and activation of oncogenes). Since REs were the sources of miRNAs in evolution, this suggests that they interact complementarily. Accordingly, REs transcripts may act as competing endogenous RNAs (ceRNAs) [133], negating the tumor suppressor effect of miRNAs. Accordingly, this is another mechanism of the activating effect of REs on oncogenes in breast cancer carcinogenesis. Thus, REs-derived microRNAs that function as tumor suppressors can be used as tools whose effect will consist not only in post-transcriptional inactivation of oncogenes, but also due to complementary interaction with REs transcripts. As a result of the introduction of high concentrations of such microRNAs, it is possible not only to eliminate the activating effect of REs transcripts on oncogenes (functioning as ceRNAs), but also to inhibit REs themselves post-transcriptionally. Accordingly, knowledge of specific REs-derived microRNAs that have an oncosuppressive effect affects the development of breast cancer in several ways (Figure 5).
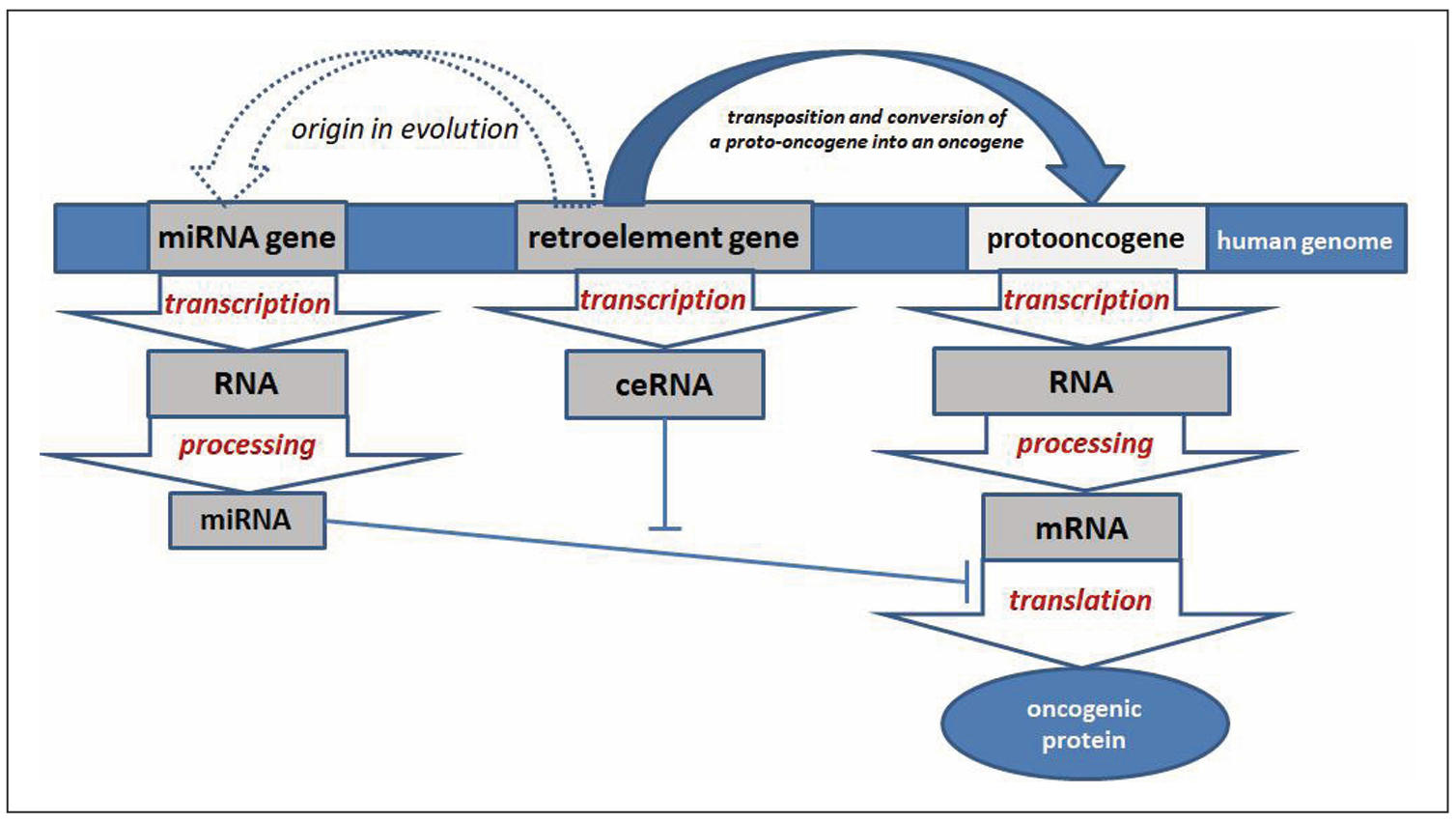
Figure 5. Scheme of interaction of retroelements with tumor suppressor microRNAs in breast cancer carcinogenesis.
Changes in microRNA levels in breast cancer are determined both in peripheral
blood and in tumor tissues. In this case, interactions of microRNA with other
ncRNAs, such as lncRNAs and circRNAs, are determined. For example, miR-28-3p
levels in the blood plasma of breast cancer patients are reduced compared to
healthy women [134]. Breast cancer tumor suppressor miR-1249-3p targeting
HOXB8 gene mRNA is a target of MIF-AS1 lncRNA that promotes breast
cancer proliferation, migration, and epithelial-mesenchymal transition [135].
MiR-28-5p, which inhibits breast cancer cell migration by regulating WSB2
(WD repeat and SOCS box contatining 2) [136], CENPF (centromere-kinetochore
complex-associated protein) [137], and LDHA (lactate dehydrogenase A) [138]
genes, is a target of lncRNA MCM3AP-AS1, which promotes breast cancer progression
[137] and circ-CSNK1G (involved in TNBC migration, invasion, and proliferation) [138].
MiR-130a-3p exhibits tumor suppressor properties by blocking the Wnt signaling
cascade in TNBC by inhibiting ID4 gene mRNA [139]. In TNBC, low levels
of miR-181c-5p are detected, which inhibits the mRNA of MAP4K4 protein (oncogenic
serine-teroneine protein kinase) [140]. MiR-330-5p, which targets mRNA of ELK1
gene, is a target of circRNA_000166, which promotes breast cancer progression [141].
MiR-342-3p functions as a tumor suppressor, inhibiting metastasis of chemoresistant
breast cancer cells by targeting ID4 mRNA (inhibits DNA-binding protein family) [142].
MiR-345-5p is a target of lncRNA RACGAP1P (a pseudogene of RACGAP1 (Ras GTPase-activating
protein 1)), which promotes metastasis and invasion of breast cancer. Interestingly,
miR-345-5p is a target of mRNA of the RACGAP1 gene, the pseudogene of which is lncRNA
that inhibits this microRNA [143].
MiR-374c-5p inhibits BC development by binding to the TATA box of the TAF7
gene [144]. MiR-422a exhibits tumor suppressor properties by inhibiting mRNA of
PLP2 gene (encodes an integral membrane protein of the endoplasmic reticulum),
preventing the formation of breast cancer microspheres and tumor cell proliferation [145].
MiR-493-5p functions as a suppressor of breast cancer cell growth and invasiveness by
targeting FUT4 gene by binding to the 3'UTR of its mRNA [146]. MiR-576-3p, a target of
oncogenic circ0012673, inhibits SOX4 (SRY-box transcription factor 4), thereby
suppressing breast cancer cell proliferation, migration, and invasion [147]. MiR-578,
a target of oncogenic circ_0008673, inhibits GINS4 (a protein that plays a role in the
initiation of DNA replication) [148]. MiR-582-3p is a negative regulator of SFXN1
gene mRNA (encodes sideroflexin, a mitochondrial membrane protein), the expression of
which is associated with poor prognosis in patients with breast cancer [149]. Expression
of tumor suppressor miR-588 is reduced in breast cancer tissues, so the use of miR-588
loaded into exosomes by electroporation, which has shown its effectiveness, can be used
in targeted therapy of breast cancer [150].
MiR-606 suppresses the development and metastasis of TNBC by affecting the STC1 gene
(encodes the glycoprotein Stannioclcin 1, which has autocrine and paracrine functions).
Transfection of miR-606 TNBC suppresses the proliferation, migration and invasion of
tumor cells, inducing their apoptosis. Therefore, this method is promising for targeted
therapy of TNBC [151]. MiR-607 suppresses the development of breast cancer by interacting
with the mRNA of EGFR gene [152]. MiR-612 inhibits myosin-2 phosphorylation and
breast cancer cell invasion [153]. MiR-625, which is the target of oncogenic breast cancer
circSEPT9, is expressed at low levels in breast cancer tissues and in breast cancer
cells. The target of miR-625 in the mechanisms of oncosuppressive action is PTBP3
(Polypyrimidine Tract Binding Protein 3), a regulator of cell differentiation [154].
MiR-634 expression is significantly reduced in breast cancer tissue and cells compared
to normal tissues. At the same time, ectopic expression of miR-634 inhibits the
proliferation and metastasis of breast cancer. The direct target of miR-634 is the
mRNA of FOXA1 gene (encodes a DNA-binding protein) [155]. MiR-640 is a
breast cancer suppressor by regulating Wnt7b/beta catenin signaling pathways [156].
MiR-646 prevents the proliferation and progression of breast cancer by inhibiting
HDAC2 (histone deacetylase) [157]. MiR-708-3p suppresses breast cancer metastases
and chemoresistance by inhibiting the epithelial-mesenchymal transition [158]. MiR-708-5p
is a target of lncRNA LOXL1-AS1, which is a driver of breast cancer cell metastasis and
invasion [159]. MiR-885-3p inhibits the PIK3/AKT pathways and NTRK2 expression, being a
target of circ_0006014, which stimulates the progression of breast cancer [160]. MiR-921,
which expression is reduced in breast cancer, inhibits the ADAM9 gene mRNA, and
is a target for circNINL [161].
MiR-1183 is the target of circ_0000851, which promotes proliferation and migration of
breast cancer cells. MiR-1183 inhibits PDK1 gene mRNA (encodes pyruvate dehydrogenase
kinase) [162]. MiR-1267 exhibits suppressive properties in relation to the initiation
and progression of breast cancer [163]. MiR-1268b expression is reduced in the tissues
of chemoresistant breast cancer compared to chemosensitive tissues [164]. MiR-1271-5p
suppresses the proliferation and progression of breast cancer by inhibiting SPIN1
gene (encodes spindylin-1, involved in the activation of Wnt pathways) [165]. MiR-1271-5p
targets the chromatin of DDIT3 gene (DNA damage-inducible transcript 3). Due to
this, repeated administration of miR-1271 effectively restores the expression levels of
alpha exogenous receptors, thereby enhancing the sensitivity of breast cancer cells to
letrozole. This is an example of the possibility of effectively using microRNAs to
eliminate breast cancer resistance to chemotherapy [166].
The worst prognosis of breast cancer in women is associated with low expression of
miR-1285-5p, which is significantly lower in all women in breast cancer samples
compared with healthy tissues. At the same time, oncosuppressive miR-1285-5p inhibits
the proliferation of breast cancer cells regardless of the tumor subtype. TMEM194
(encodes the transmembrane protein 194A) plays an important role among the target genes
of this microRNA [167]. MiR-1294 suppresses the development of breast cancer by regulating
ERK signaling [168]. MiR-1303 inhibits the PTBP3 gene, a protein product of which
stimulates the development of breast cancer. Accordingly, lncRNA BCRT1, which binds
complementarily to miR-1303, is oncogenic for breast cancer [169]. Oncosuppressive miR-1972
can be used for effective targeted breast cancer therapy, since the introduction of miR-1972
mimics inhibits genes involved in angiogenesis and metastasis (SHC1, NEK1, MMP9, MAP2K1),
suppressing the proliferation of breast cancer cells [170]. MiR-2355-5p is a target of lncRNA
SNHG11, which promotes proliferation and migration of breast cancer cells. MiR-2355-5p targets
the mRNA of the CBX5 gene (encodes a non-histone protein, a member of the heterochromatin
protein complex) [171].
Reduced levels of miR-3139, which is a target for LINC00514, were detected in breast
cancer tissues (this dnRNA promotes proliferation, migration, and invasion of breast
cancer by suppressing apoptosis) [172]. MiR-3163 expression, which functions as a tumor
suppressor, is reduced in breast cancer tissues compared to normal tissue [173]. MiR-3194-3p
inhibits the progression of breast cancer by interacting with the mRNA of the AQP1
gene (encodes aquaporin 1) [174]. Significant increase in the expression of miR-3674, which
targets mRNA of ATF3 (encodes transcription factor) gene, was detected in breast
cancer cells [175]. The association of low miR-3923 expression with lymph node metastases
in patients with breast cancer has been determined [176]. Oncogenic miR-4472, whose expression
is significantly increased in breast cancer metastatic tissues, promotes breast cancer
proliferation and aggressiveness by targeting RGMA (encodes Repulsive guidance
molecule A, cancer metastasis suppressor) and inducing the epithelial-mesenchymal transition
by suppressing E-cadherin with the initiation of vimentin, Slug and beta-catenin [177]. Breast
cancer is characterized by decreased expression of oncosuppressive miR-5586-5p, which inhibits
angiogenesis by suppressing VEGFA (encodes vascular endothelial growth factor A),
ANGPTL4 (encodes angiopoietin-like protein 4), HBEGF (encodes heparin-binding
EGF-like factor) genes [178]. MiR-7978, which is the target of oncogenic BC circHSDL2, inhibits
ZNF704, thus regulating Hippo pathways [179].
Among the 43 microRNAs that originated from REs, the expression of which is reduced during
breast cancer, 13 microRNAs have a change in expression with aging. At the same time, 5
microRNAs (miR-28-3p, miR-330-5p, miR-493-5p, miR-582-3p, miR-2355-5p) showed increased
levels in various tissues during aging. This indicates that not all epigenetic mechanisms of
aging involving oncosuppressive microRNAs derived from REs play a role in the initiation and
development of breast cancer. This circumstance is logical, since not all women develop breast
cancer with age. Changes in the expression of such microRNAs have been detected in various
organs and tissues. For example, during early replicative senescence of endothelial cells,
a significant increase in miR-28-3p expression was detected [180]. An increase in the expression
of miR-330-5p was detected during aging of bone marrow mesenchymal stem cells [181]. Accumulation
of kynurenine, a metabolite of tryptophan, plays a role in age-related pathophysiological changes,
including bone loss. As a result of this accumulation, proliferation and differentiation of
bone marrow stromal cells decrease. In experiments on human bone marrow stromal cells treated
with kynurenine, miR-493-5p expression increased [182]. A decrease in miR-493-5p expression
was also detected in senescent pig skeletal muscle cells [183]. Increased levels of miR-582-3p
in liver tissue during aging have been determined [184]. Increased levels of miR-2355-5p have
been determined by inhibiting ERRFI1 during aging and intervertebral disc
degeneration [185].
For 8 microRNAs derived from REs, the expression of which is reduced during breast cancer,
a decrease in levels during aging is also determined: miR-28-5p, miR-181c-5p, miR-588, miR-612,
miR-708-5p, miR-1249-3p, miR-1271-5p, miR-1972. This indicates that the epigenetic mechanisms
of aging are important in the development of breast cancer. A decrease in the expression of
miR-28-5p in human blood plasma during physiological aging was revealed [186]. In blood
plasma, a decrease in miR-181c-5p expression with age correlates with the accumulation
of amyloid beta [187]. An increase in the expression of miR-1249-3p in the ovaries during
aging has been determined [188]. Increased expression of circRNAs was detected in aging
skin samples: circRNA-0088179, -0132428, -0094423, -0008166, -0138184, -0135743, -0114119,
-0131421, the targets of which are miR-588 and miR-612. Accordingly, the levels of these
microRNAs decrease with skin aging [189]. During aging, a decrease in miR-708-5p expression
is detected in various tissues and cells. MiR-708-5p directly inhibits the mRNA of the
Dab2 gene, as a result of which the activation of mTORC1 is suppressed. In addition,
activation of AMP-activating protein kinase stimulates miR-708-5p by increasing DICER
expression [190]. In human mesenchymal stromal cells, a decrease in miR-1271-5p levels
was detected during their aging [191]. In elderly people with physiological aging,
leukoarrhoea of the brain is observed in the form of hyperintensivity, in which there is
a decrease in the expression of miR-1972, the target of which is the mRNA of BAIAP3
gene [192]. Table 2 shows oncosuppressive microRNAs
derived from retroelements and changes in their activity during aging. The use of the
above-mentioned oncosuppressive microRNAs as tools will not only suppress the functions of
oncogenes, but also competitively inhibit REs transcripts, which is associated with the
complementarity of the sequences of such microRNAs with the REs from which they originated.
Taking into account the role of such microRNAs in aging, a differentiated approach to the
methods of administration is possible, since it is most advisable for microRNAs that promote
aging to be injected directly into the tumor tissue.
Table 2.
ERetroelement-derived microRNAs which expression is decreased in breast cancer and altered with aging.
| RE-source of microRNA | microRNA/ tissue | miRNA target genes/interaction with other ncRNAs | changes with aging (tissue) | |
|---|---|---|---|---|
| 1 | LINE2 | miR-28-5p/ blood plasma [136-138] | WSB2 [136], CENPF [137], LDHA/ lncRNA MCM3AP-AS1 [137], circ-CSNK1G [138] | decreased (human blood plasma) [186] |
| 2 | LINE-RTE-BovB | miR-181c-5p/ cancer [140] | MAP4K4 [140] | decreased (human blood plasma) [187] |
| 3 | LINE1 | miR-588/ cancer [150] | SPTLC2, RHBDF1, MGAT1, PRKAR1A, COL4A4, TUSC1, RIMS1 (miRDB) | decreased (skin tissue) [189] |
| 4 | SINE-MIR | miR-612/ cancer [153] | AH1, BTRC, DAB2IP, FMO5, SIKE1, SAR1A, SEC63 (miRDB) | decreased (human skin) [189] |
| 5 | LINE2 | miR-708-5p/ cancer [159] | TNS3, FOXJ3, IKBKB, ASPA, BCAM, NPY2R, ARL17A, CNTFR (miRDB) / lncRNA LOXL1-AS1 [159] | decreased (human skin) [190] |
| 6 | LINE2 | miR-1249-3p/ cancer [135] | HOXB8 [135] | decreased (ovaries) [188] |
| 7 | LINE2 | miR-1271-5p/ cancer [165, 166] | DDIT3 [166], SPIN1 [165] | decreased (human mesenchymal stromal cells) [191] |
| 8 | SINE-Alu | miR-1972/ cancer [170] | SHC1, NEK1, MMP9, MAP2K1 [170], BAIAP3 [192] | decreased (brain) [192] |
| 9 | LINE2 | miR-28-3p/ blood plasma [134] | FRMD7, VIM, RSBN1L, C5, MBL2, RAD51B, UNKL, IKZF5 (miRDB) | increased (human endothelial cell culture) [180] |
| 10 | SINE-MIR | miR-330-5p/ cancer [141] | ELK1/ circRNA_000166 [141] | increased (bone marrow mesenchymal stem cells) [181] |
| 11 | LINE2 | miR-493-5p/ cancer [146] | FUT4 [146], WNT5A, SOX1 [182] | increased (human bone marrow stromal cells) [182] |
| 12 | LINE-CR1 | miR-582-3p/ cancer [149] | SFXN1 [149] | increased (liver tissue) [184] |
| 13 | LINE-RTE-BovB | miR-2355-5p/ cancer [171] | ERRFI1 [185], CBX5/ lncRNA SNHG11 [171] | increased (cartilage tissue) [185] |
| 14 | LINE-RTE-BovB | miR-130a-3p/ cancer [139] | ID4 [139] | data not found |
| 15 | SINE-tRNA-RTE | miR-342-3p/ cancer [142] | ID4 [142] | data not found |
| 16 | SINE-MIR | miR-345-5p/ cancer, breast cancer cell line [143] | RACGAP1/ lncRNA RACGAP1P [143] | data not found |
| 17 | LINE2 | MiR-374c-5p/ cancer [144] | TAF7 [144] | data not found |
| 18 | SINE-MIR | miR-422a/ cancer, breast cancer cell line [145] | PLP2 [145] | data not found |
| 19 | LINE1 | miR-576-3p/ cancer [147] | SOX4/ circ0012673 [147] | data not found |
| 20 | LINE2 | miR-578/ cancer [148] | GINS4/ circ_0008673 [148] | data not found |
| 21 | LINE1 | miR-606/ cancer [151] | STC1 [151] | data not found |
| 22 | SINE-MIR | miR-607/ cancer [152] | EGFR [152] | data not found |
| 23 | LINE1 | miR-625-5p/ cancer [154] | PTBP3/ circSEPT9 [154] | data not found |
| 24 | LINE1 | miR-634/ cancer [155] | FOXA1 [155] | data not found |
| 25 | SINE-MIR | miR-640/ cancer [156] | Wnt7b [156] | data not found |
| 26 | ERVL | miR-646/ cancer [157] | HDAC2 [157] | data not found |
| 27 | LINE2 | miR-708-3p/ cancer, breast cancer cell line [158] | SCAMP1, JARID2, TRAF3, VIM, BAZ1B, SPRED1, HSPA4L (miRDB) | data not found |
| 28 | SINE-MIR | miR-885-3p/ cancer [160] | NTRK2/ circ_0006014 [160] | data not found |
| 29 | SINE-MIR | miR-921/ cancer [161] | ADAM9/ circNINL [161] | data not found |
| 30 | LINE2 | miR-1183/ cancer [162] | PDK1/ circ_0000851 [162] | data not found |
| 31 | ERVL-MaLR | miR-1267/ cancer [163] | SALL1, SMIM8, MAN2A1, RUFY2, SHROOM2, BBS10, DDI2 (miRDB) | data not found |
| 32 | Alu | miR-1268b/ cancer [164] | STXBP1, EEPD1, EGR4, CMIP FCAR, HPD, USB1, SLC8A2 (miRDB) | data not found |
| 33 | Alu | miR-1285-5p/ cancer [167] | TMEM194 [167] | data not found |
| 34 | SINE-MIR | miR-1294/ cancer [168] | PAPPA, INPP5A, CD164, MYCL, SKIL, CDC34, THRSP, HAS2 (miRDB) | data not found |
| 35 | Alu | miR-1303/ cancer [169] | PTBP3/ lncRNA BCRT1 [169] | data not found |
| 36 | LINE2 | miR-3139/ cancer [172] | TNS3, IKBKB, FOXJ3, ARL17A, RPTN (miRDB)/ LINC00514 [172] | data not found |
| 37 | SINE | miR-3163/ cancer [173] | SMC3, AHCTF1, HCN1, MEI4, PDK3, ZC3H6, TMX3, HIPK3, SAMD8 (miRDB) | data not found |
| 38 | SINE-MIR | miR-3193-3p/ cancer [175] | AQP1 [174] | data not found |
| 39 | LTR-ERV1 | miR-3674/ cancer, breast cancer cells [175] | ATF3 [175] | data not found |
| 40 | ERVL-MaLR | miR-3923/ cancer [176] | MSR1, HDX, ST8SIA1, CPED1 (miRDB) | data not found |
| 41 | Alu | miR-4472/ cancer [177] | RGMA, CDH1 [177] | data not found |
| 42 | LINE1 | miR-5586-5p/ breast cancer cells [178] | ANGPTL4, VEGFA, HBEGF [178] | data not found |
| 43 | LINE1 | MiR-7978/ cancer [179] | ZNF704/ circHSDL2 [179] | data not found |
Summarizing the overall trends in Table 2, it can be seen that among the 43 REs-derived miRNAs whose levels are reduced in BC, 8 miRNAs (of which 6 miRNAs are LINE-derived and 2 miRNAs are SINE-derived) also show a decrease in expression with aging. This may indicate that the downregulation of retroelement-derived tumor suppressor miRNAs during aging is a trigger for the development and progression of breast cancer. REs (from which such tumor suppressor miRNAs originate) activated in such pathways contribute to the downregulation of these miRNAs by complementary binding of their transcripts, acting as ceRNAs [133]. This may explain the decrease in expression of tumor suppressor microRNAs derived from REs due to activation of such REs. In addition, Table 2 presents 5 microRNAs derived from REs, the expression of which is decreased in breast cancer but increased in aging. This circumstance may be explained by the fact that not all molecular epigenetic mechanisms of aging contribute to breast cancer carcinogenesis, but many of them do (8 microRNAs are more than 5 microRNAs). In addition, for 30 microRNAs presented in Table 2, no evidence of involvement of these microRNAs in aging mechanisms was found in the scientific literature. All this suggests that breast cancer carcinogenesis is a complex multicomponent process in which aging and REs activation are some of the links in the development of pathology, but are not the only factors in breast cancer carcinogenesis. However, research in this area can become the basis for more effective therapy of the disease and for explaining individual components of breast cancer pathogenesis. The RE-derived tumor suppressor miRNAs described in Table 2 can be used as tools for specific inhibition of retroelements and oncogenes that are pathologically activated in breast cancer. Thus, the use of such miRNAs can have a dual-targeted antitumor effect in the treatment of breast cancer. An additional mechanism of action on activated REs in these processes may be the use of reverse transcriptase inhibitors [75, 76].
Conclusions
TAnalysis of scientific literature indicates the role of epigenetic mechanisms of aging in the development of breast cancer as a multifactorial disease. In these mechanisms, during aging, epigenetic derepression of retroelements occurs, the activation of which is a factor in carcinogenesis due to the inactivation of tumor suppressor genes, the induction of genomic instability and the activation of oncogenes. Breast cancer-associated SNPs may influence changes in the functioning and activation of retroelements, potentiating age-associated breast cancer development. This may explain why not all women develop breast cancer with aging. In addition, breast cancer-associated SNPs may alter the nucleotide sequences of retroelements, disrupting their complementary interaction with microRNAs, since retroelement transcripts function as ceRNAs. Accordingly, changes in retroelement activity in breast cancer are reflected in the levels of microRNAs derived from them. Indeed, analysis of the MDTE database with scientific literature data on the effect of specific microRNAs on breast cancer development revealed a decrease in the expression of 43 microRNAs derived from retroelements and an increase in the levels of 43 microRNAs derived from retroelements in breast cancer. Moreover, 30 of these miRNAs are characterized by impaired expression with aging, which indicates epigenetic relationships at the level of complementary interactions of retroelements with miRNAs between aging and breast cancer development. The use of the obtained data in modern medicine for targeted therapy of breast cancer using the described microRNAs to modulate the activity of retroelements is promising. The use of retroelement-derived microRNAs described in the article is promising for targeted therapy of breast cancer in personalized medicine, since they can be used to influence specific molecular links in breast cancer carcinogenesis. Namely, onosuppressive microRNAs can be used as tools for suppressing the expression of REs (due to complementary interaction with their transcripts) and oncogenes. In these pathways, the use of such microRNAs as miR-28-5p, miR-181c-5p, miR-588, miR-612, miR-708-5p, miR-1249-3p, miR-1271-5p, miR-1972 can also slow down the aging process, since the levels of these microRNAs are reduced during aging. Oncogenic microRNAs derived from REs can be used as targets for their inhibition using antisense oligonucleotides. This method can also inhibit pathologically activated REs in the treatment of breast cancer through complementary nucleotide binding. Moreover, the use of antisense nucleotides against miR-378d, miR-552-5p, miR-558, miR-576-5p, miR-1272-3p, miR-3135b, miR-4428, miR-4480, miR-5100 can also have an anti-aging effect, since the expression of these microRNAs decreases with age.
Declarations
Author contributions
The author contributed solely to the article.
Availability of data and materials
Not applicable.
Financial support and sponsorship
None.
Conflicts of interests
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
References
1. Mak J, McMurran C, Kuja-Halkola R, Hall P, Czene K, Jylhävä J, et al. Clinical biomarker-based biological aging and risk of cancer in the UK Biobank. Br J Cancer, 2023, 129(1): 94-103. [Crossref]
2. Bai H, Liu X, Lin M, Meng Y, Tang R, Guo Y, et al. Progressive senescence programs induce intrinsic vulnerability to aging-related female breast cancer. Nature Communications, 2024, 15(1): 5154-5164. [Crossref]
3. Chang L, Weiner L, Hartman S, Horvath S, Jeste D, Mischel P, et al. Breast cancer treatment and its effects on aging. J Geriatr Oncol, 2019, 10(2): 346-355. [Crossref]
4. Zhu J, Wang F, Shi L, Cai H, Zheng Y, Zheng W, et al. Accelerated aging in breast cancer survivors and its association with mortality and cancer recurrence. Breast Cancer Res Treat, 2020, 180(2): 449-459. [Crossref]
5. Sokolova A, Johnstone K, McCart Reed A, Simpson P, & Lakhani S. Hereditary breast cancer: syndromes, tumour pathology and molecular testing. Histopathology, 2023, 82(1): 70-82. [Crossref]
6. Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush M, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet, 2015, 47(4): 373-380. [Crossref]
7. Zhang H, Ahearn T, Lecarpentier J, Barnes D, Beesley J, Qi G, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat Genet, 2020, 52(6): 572-581. [Crossref]
8. Roberts E, Howell S, & Evans D. Polygenic risk scores and breast cancer risk prediction. Breast, 2023, 67: 71-77. [Crossref]
9. Sherman M, Winham S, Vierkant R, McCauley B, Scott C, Schrup S, et al. Polygenic risk scores stratify breast cancer risk among women with benign breast disease. J Natl Cancer Inst, 2025, 117(3): 456-464. [Crossref]
10. Sun S, Yin S, Huang J, Zhou D, Tan Q, Man X, et al. Identification of significant single-nucleotide polymorphisms associated with breast cancer recurrence and metastasis using GWAS. Cancer Innov, 2025, 4(1): e142. [Crossref]
11. Jia G, Chen Z, Ping J, Cai Q, Tao R, Li C, et al. Refining breast cancer genetic risk and biology through multi-ancestry fine-mapping analyses of 192 risk regions. Nat Genet, 2025, 57(1): 80-87. [Crossref]
12. Yong S, Raben T, Lello L, & Hsu S. Genetic architecture of complex traits and disease risk predictors. Sci Rep, 2020, 10(1): 12055. [Crossref]
13. Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, et al. The complete sequence of a human genome. Science, 2022, 376(6588): 44-53. [Crossref]
14. RN M. The role of transposable elements in the association of polymorphic variants with multifactorial diseases. Opera Medica et Physiologica, 2024, 11(4): 60-70.
15. Park E, Ha H, Lee D, Kim W, Lee Y, Bae W, et al. Genomic analyses of non-coding rnas overlapping transposable elements and its implication to human diseases. Int J Mol Sci, 2022, 23(16): 8950-8960. [Crossref]
16. Zabihi N, Sadeghi S, Tabatabaeian H, Ghaedi K, Azadeh M, & Fazilati M. The association between rs1972820 and the risk of breast cancer in Isfahan population. J Cancer Res Ther, 2017, 13(1): 26-32. [Crossref]
17. Rybarczyk A, Lehmann T, Iwańczyk-Skalska E, Juzwa W, Pławski A, Kopciuch K, et al. In silico and in vitro analysis of the impact of single substitutions within EXO-motifs on Hsa-MiR-1246 intercellular transfer in breast cancer cell. J Appl Genet, 2023, 64(1): 105-124. [Crossref]
18. Dai Y, Pan Y, Quan M, Chen Q, Pan Y, Ruan Y, et al. MicroRNA-1246 mediates drug resistance and metastasis in breast cancer by targeting NFE2L3. Front Oncol, 2021, 11: 677168. [Crossref]
19. Gorbunova V, Seluanov A, Mita P, McKerrow W, Fenyö D, Boeke J, et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature, 2021, 596(7870): 43-53. [Crossref]
20. Kankava K, Kvaratskhelia E, Burkadze G, Kokhreidze I, Gogokhia N, & Abzianidze E. LINE-1 methylation in blood and tissues of patients with breast cancer. Georgian Med News, 2018, (276): 107-112.
21. van Hoesel A, van de Velde C, Kuppen P, Liefers G, Putter H, Sato Y, et al. Hypomethylation of LINE-1 in primary tumor has poor prognosis in young breast cancer patients: a retrospective cohort study. Breast Cancer Res Treat, 2012, 134(3): 1103-1114. [Crossref]
22. Park S, Seo A, Jung H, Gwak J, Jung N, Cho N, et al. Alu and LINE-1 hypomethylation is associated with HER2 enriched subtype of breast cancer. PLoS One, 2014, 9(6): e100429. [Crossref]
23. Jiang J, Rothnagel J, & Upton K. Widespread exaptation of L1 transposons for transcription factor binding in breast cancer. Int J Mol Sci, 2021, 22(11): 5625-5635. [Crossref]
24. Berrino E, Miglio U, Bellomo S, Debernardi C, Bragoni A, Petrelli A, et al. The tumor-specific expression of L1 retrotransposons independently correlates with time to relapse in hormone-negative breast cancer patients. Cells, 2022, 11(12): 1944-1954. [Crossref]
25. Jang H, Shah N, Du A, Dailey Z, Pehrsson E, Godoy P, et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nature Genetics, 2019, 51(4): 611-617. [Crossref]
26. Rodriguez-Martin B, Alvarez E, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet, 2020, 52(3): 306-319. [Crossref]
27. Ramos K, Montoya-Durango D, Teneng I, Nanez A, & Stribinskis V. Epigenetic control of embryonic renal cell differentiation by L1 retrotransposon. Birth Defects Res A Clin Mol Teratol, 2011, 91(8): 693-702. [Crossref]
28. Shukla R, Upton K, Muñoz-Lopez M, Gerhardt D, Fisher M, Nguyen T, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell, 2013, 153(1): 101-111. [Crossref]
29. Xia Z, Cochrane D, Anglesio M, Wang Y, Nazeran T, Tessier-Cloutier B, et al. LINE-1 retrotransposon-mediated DNA transductions in endometriosis associated ovarian cancers. Gynecol Oncol, 2017, 147(3): 642-647. [Crossref]
30. Cajuso T, Sulo P, Tanskanen T, Katainen R, Taira A, Hänninen U, et al. Retrotransposon insertions can initiate colorectal cancer and are associated with poor survival. Nat Commun, 2019, 10(1): 4022-4032. [Crossref]
31. Aschacher T, Wolf B, Enzmann F, Kienzl P, Messner B, Sampl S, et al. LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene, 2016, 35(1): 94-104. [Crossref]
32. Nair M, Ramesh R, Naidu C, Mavatkar A, V P, Ramamurthy V, et al. Estimation of ALU repetitive elements in plasma as a cost-effective liquid biopsy tool for disease prognosis in breast cancer. Cancers, 2023, 15(4): 1054-1055. [Crossref]
33. RN M. Participation of retroelements in chromoanagenesis in cancer development. Siberian journal of oncology, 2024, 23(5): 146-156. [Crossref]
34. Bolkestein M, Wong J, Thewes V, Körber V, Hlevnjak M, Elgaafary S, et al. Chromothripsis in human breast cancer. Cancer Res, 2020, 80(22): 4918-4931. [Crossref]
35. Vasmatzis G, Wang X, Smadbeck J, Murphy S, Geiersbach K, Johnson S, et al. Chromoanasynthesis is a common mechanism that leads to ERBB2 amplifications in a cohort of early stage HER2+ breast cancer samples. BMC Cancer, 2018, 18(1): 738-743. [Crossref]
36. Fazza A, Sabino F, de Setta N, Bordin N, Jr., da Silva E, & Carareto C. Estimating genomic instability mediated by Alu retroelements in breast cancer. Genet Mol Biol, 2009, 32(1): 25-31. [Crossref]
37. Chisholm K, Aubert S, Freese K, Zakian V, King M, & Welcsh P. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS One, 2012, 7(2): e30748. [Crossref]
38. Denariyakoon S, Puttipanyalears C, Chatamra K, & Mutirangura A. Breast cancer sera changes in Alu element methylation predict metastatic disease progression. Cancer Diagn Progn, 2022, 2(6): 731-738. [Crossref]
39. Chen Y, Salas L, Marotti J, Jenkins N, Cheng C, Miller T, et al. Extensive epigenomic dysregulation is a hallmark of homologous recombination deficiency in triple-negative breast cancer. Int J Cancer, 2025, 156(6): 1191-1202. [Crossref]
40. Gelaleti G, Granzotto A, Leonel C, Jardim B, Moschetta M, Carareto C, et al. Short interspersed CAN SINE elements as prognostic markers in canine mammary neoplasia. Oncol Rep, 2014, 31(1): 435-441. [Crossref]
41. Park J, Kim H, Yun S, & Kim S. Ginsenoside Rh2 upregulates long noncoding RNA STXBP5-AS1 to sponge microRNA-4425 in suppressing breast cancer cell proliferation. J Ginseng Res, 2021, 45(6): 754-762. [Crossref]
42. Abd El Hafeez H, Abd El Rahman M, Kamel T, Rezk K, Mohamed F, & Abdel-Hameed Z. The role of circulating cell-free DNA and its integrity as a biomarker for diagnosis of breast cancer using ALU (247/115) bP sequences. Egypt J Immunol, 2023, 30(3): 44-55.
43. Özgür E, Ferhatoğlu F, Şen F, & Gezer U. Effect of cytotoxic Therapy on ALU expression is modulated by hormonal status in patients with breast cancer. Balkan Med J, 2022, 39(6): 450-451. [Crossref]
44. Özgür E, Ferhatoglu F, Sen F, Saip P, & Gezer U. Expression of Alu repeat in blood plasma of patients with breast cancer during neoadjuvant chemotherapy: an exploratory study. Exp Oncol, 2023, 45(1): 120-124. [Crossref]
45. Song R, Guo P, Ren X, Zhou L, Li P, Rahman NA, et al. A novel polypeptide CAPG-171aa encoded by circCAPG plays a critical role in triple-negative breast cancer. Mol Cancer, 2023, 22(1): 104-115. [Crossref]
46. Wang X, Chen T, Li C, Li W, Zhou X, Li Y, et al. CircRNA-CREIT inhibits stress granule assembly and overcomes doxorubicin resistance in TNBC by destabilizing PKR. J Hematol Oncol, 2022, 15(1): 122-134. [Crossref]
47. Wu Q, Nie D, Ba-Alawi W, Ji Y, Zhang Z, Cruickshank J, et al. PRMT inhibition induces a viral mimicry response in triple-negative breast cancer. Nat Chem Biol, 2022, 18(8): 821-830. [Crossref]
48. Cheng J, Cuk K, Heil J, Golatta M, Schott S, Sohn C, et al. Cell-free circulating DNA integrity is an independent predictor of impending breast cancer recurrence. Oncotarget, 2017, 8(33): 54537-54547. [Crossref]
49. Annapragada A, Niknafs N, White J, Bruhm D, Cherry C, Medina J, et al. Genome-wide repeat landscapes in cancer and cell-free DNA. Sci Transl Med, 2024, 16(738): eadj9283. [Crossref]
50. Ono M, Kawakami M, & Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol, 1987, 61(6): 2059-2062. [Crossref]
51. Wang-Johanning F, Frost A, Johanning G, Khazaeli M, LoBuglio A, Shaw D, et al. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res, 2001, 7(6): 1553-1560.
52. Wang-Johanning F, Frost A, Jian B, Epp L, Lu D, & Johanning G. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene, 2003, 22(10): 1528-1535. [Crossref]
53. Nguyen T, Davis J, Eugenio R, & Liu Y. Female sex hormones activate human endogenous retrovirus type k through the Oct4 transcription factor in T47D breast cancer cells. AIDS Res Hum Retroviruses, 2019, 35(3): 348-356. [Crossref]
54. Zhou F, Li M, Wei Y, Lin K, Lu Y, Shen J, et al. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget, 2016, 7(51): 84093-84117. [Crossref]
55. Wang T, Zeng J, Lowe C, Sellers R, Salama S, Yang M, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA, 2007, 104(47): 18613-18618. [Crossref]
56. Pollock N, Ramroop J, Hampel H, Troester M, Conway K, Hu J, et al. Differences in somatic TP53 mutation type in breast tumors by race and receptor status. Breast Cancer Res Treat, 2022, 192(3): 639-648. [Crossref]
57. Arancio W, & Coronnello C. Repetitive sequence transcription in breast cancer. Cells, 2022, 11(16): 2522-2534. [Crossref]
58. Lu X, Sachs F, Ramsay L, Jacques P, Göke J, Bourque G, et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol, 2014, 21(4): 423-425. [Crossref]
59. Rivas S, Valdez M, Govindarajan V, Seetharam D, Doucet-O'Hare T, Heiss J, et al. The role of HERV-K in cancer stemness. Viruses, 2022, 14(9): 2019-2024. [Crossref]
60. Mustafin R. The relationship between transposons and transcription factors in the evolution of eukaryotes. Journal of Evolutionary Biochemistry and Physiology, 2019, 55(1): 14-23. [Crossref]
61. Babaian A, Romanish M, Gagnier L, Kuo L, Karimi M, Steidl C, et al. Onco-exaptation of an endogenous retroviral LTR drives IRF5 expression in Hodgkin lymphoma. Oncogene, 2016, 35(19): 2542-2546. [Crossref]
62. Lock F, Rebollo R, Miceli-Royer K, Gagnier L, Kuah S, Babaian A, et al. Distinct isoform of FABP7 revealed by screening for retroelement-activated genes in diffuse large B-cell lymphoma. Proc Natl Acad Sci USA, 2014, 111(34): E3534-3543. [Crossref]
63. Scarfò I, Pellegrino E, Mereu E, Kwee I, Agnelli L, Bergaggio E, et al. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood, 2016, 127(2): 221-232. [Crossref]
64. Wiesner T, Lee W, Obenauf A, Ran L, Murali R, Zhang Q, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature, 2015, 526(7573): 453-457. [Crossref]
65. Bouras A, Leone M, Bonadona V, Lebrun M, Calender A, & Boutry-Kryza N. Identification and characterization of new Alu element insertion in the BRCA1 Exon 14 associated with hereditary breast and ovarian cancer. Genes, 2021, 12(11): 1736-1743. [Crossref]
66. RN. M, & EK K. The role of retroelements in the development of hereditary tumor syndromes. Advances in Molecular Oncology, 2021, 8(4): 42-52.
67. Ji X, & Zhao S. DA and Xiao—two giant and composite LTR–retrotransposon-like elements identified in the human genome. Genomics, 2008, 91(3): 249-258. [Crossref]
68. Liang B, Yan T, Wei H, Zhang D, Li L, Liu Z, et al. HERVK-mediated regulation of neighboring genes: implications for breast cancer prognosis. Retrovirology, 2024, 21(1): 4-15. [Crossref]
69. Lemaître C, Tsang J, Bireau C, Heidmann T, & Dewannieux M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog, 2017, 13(6): e1006451. [Crossref]
70. Rhyu D, Kang Y, Ock M, Eo J, Choi Y, Kim W, et al. Expression of human endogenous retrovirus env genes in the blood of breast cancer patients. Int J Mol Sci, 2014, 15(6): 9173-9183. [Crossref]
71. Johanning G, Malouf G, Zheng X, Esteva F, Weinstein J, Wang-Johanning F, et al. Expression of human endogenous retrovirus-K is strongly associated with the basal-like breast cancer phenotype. Sci Rep, 2017, 7: 41960. [Crossref]
72. Wang-Johanning F, Li M, Esteva F, Hess K, Yin B, Rycaj K, et al. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int J Cancer, 2014, 134(3): 587-595. [Crossref]
73. de Koning A, Gu W, Castoe T, Batzer M, & Pollock D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet, 2011, 7(12): e1002384. [Crossref]
74. Okumura S, Hirano Y, & Komatsu Y. Inhibition of breast cancer cell proliferation with anti-microRNA oligonucleotides flanked by interstrand cross-linked duplexes. Nucleosides Nucleotides Nucleic Acids, 2020, 39(1-3): 225-235. [Crossref]
75. Patnala R, Lee S, Dahlstrom J, Ohms S, Chen L, Dheen S, et al. Inhibition of LINE-1 retrotransposon-encoded reverse transcriptase modulates the expression of cell differentiation genes in breast cancer cells. Breast Cancer Res Treat, 2014, 143(2): 239-253. [Crossref]
76. Şekeroğlu Z, Şekeroğlu V, & Küçük N. Effects of reverse transcriptase inhibitors on proliferation, apoptosis, and migration in breast carcinoma cells. Int J Toxicol, 2021, 40(1): 52-61. [Crossref]
77. Yang Q, Zhao S, Shi Z, Cao L, Liu J, Pan T, et al. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J Exp Clin Cancer Res, 2021, 40(1): 120-131. [Crossref]
78. Sun T, Li Y, Zhao F, Sun H, Gao Y, Wu B, et al. MiR-1-3p and MiR-124-3p synergistically damage the intestinal barrier in the ageing colon. J Crohns Colitis, 2022, 16(4): 656-667. [Crossref]
79. Jiang B, Xia J, & Zhou X. Overexpression of lncRNA SLC16A1-AS1 suppresses the growth and metastasis of breast cancer via the miR-552-5p/WIF1 signaling pathway. Front Oncol, 2022, 12: 712475. [Crossref]
80. Breunig S, Wallner V, Kobler K, Wimmer H, Steinbacher P, Streubel M, et al. The life in a gradient: calcium, the lncRNA SPRR2C and mir542/mir196a meet in the epidermis to regulate the aging process. Aging, 2021, 13(15): 19127-19144. [Crossref]
81. Zhu H, Dai M, Chen X, Chen X, Qin S, & Dai S. Integrated analysis of the potential roles of miRNA‑mRNA networks in triple negative breast cancer. Mol Med Rep, 2017, 16(2): 1139-1146. [Crossref]
82. Yu J, Wu X, Gimble J, Guan X, Freitas M, & Bunnell B. Age-related changes in mesenchymal stem cells derived from rhesus macaque bone marrow. Aging cell, 2011, 10(1): 66-79. [Crossref]
83. Li D, & Hu A. LINC-PINT suppresses breast cancer cell proliferation and migration via MEIS2/PPP3CC/NF-κB pathway by sponging miR-576-5p. Am J Med Sci, 2024, 367(3): 201-211. [Crossref]
84. Ipson B, Fletcher M, Espinoza S, & Fisher A. Identifying exosome-derived microRNAs as candidate biomarkers of frailty. J Frailty Aging, 2018, 7(2): 100-103. [Crossref]
85. Turkistani S, Sugita B, Fadda P, Marchi R, Afsari A, Naab T, et al. A panel of miRNAs as prognostic markers for African-American patients with triple negative breast cancer. BMC Cancer, 2021, 21(1): 861-873. [Crossref]
86. Somel M, Guo S, Fu N, Yan Z, Hu H, Xu Y, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res, 2010, 20(9): 1207-1218. [Crossref]
87. Koleckova M, Ehrmann J, Bouchal J, Janikova M, Brisudova A, Srovnal J, et al. Epithelial to mesenchymal transition and microRNA expression are associated with spindle and apocrine cell morphology in triple-negative breast cancer. Sci Rep, 2021, 11(1): 5145-5155. [Crossref]
88. Alberro A, Bravo-Miana R, Gs Iñiguez S, Iribarren-López A, Arroyo-Izaga M, Matheu A, et al. Age-related sncRNAs in human hippocampal tissue samples: focusing on deregulated miRNAs. Int J Mol Sci, 2024, 25(23): 12872. [Crossref]
89. Sato J, Shimomura A, Kawauchi J, Matsuzaki J, Yamamoto Y, Takizawa S, et al. Brain metastasis-related microRNAs in patients with advanced breast cancer. PLoS One, 2019, 14(10): e0221538. [Crossref]
90. Machida T, Tomofuji T, Ekuni D, Maruyama T, Yoneda T, Kawabata Y, et al. MicroRNAs in salivary exosome as potential biomarkers of aging. Int J Mol Sci, 2015, 16(9): 21294-21309. [Crossref]
91. Popp N, Yu D, Green B, Chew E, Ning B, Chan C, et al. Functional single nucleotide polymorphism in IL-17A 3' untranslated region is targeted by miR-4480 in vitro and may be associated with age-related macular degeneration. Environ Mol Mutagen, 2016, 57(1): 58-64. [Crossref]
92. Sathipati S, Tsai M, Aimalla N, Moat L, Shukla S, Allaire P, et al. An evolutionary learning-based method for identifying a circulating miRNA signature for breast cancer diagnosis prediction. NAR Genom Bioinform, 2024, 6(1): lqae022.
93. Engel A, Wagner V, Hahn O, Foltz A, Atkins M, Beganovic A, et al. A spatio-temporal brain miRNA expression atlas identifies sex-independent age-related microglial driven miR-155-5p increase. bioRxiv, 2025, 643430. [Crossref]
94. Wang Y, Yin W, Lin Y, Yin K, Zhou L, Du Y, et al. Downregulated circulating microRNAs after surgery: potential noninvasive biomarkers for diagnosis and prognosis of early breast cancer. Cell Death Discov, 2018, 4: 21-33. [Crossref]
95. Noren Hooten N, Fitzpatrick M, Wood W, 3rd, De S, Ejiogu N, Zhang Y, et al. Age-related changes in microRNA levels in serum. Aging, 2013, 5(10): 725-740. [Crossref]
96. Wang H, Hu X, Yang F, & Xiao H. miR-325-3p promotes the proliferation, invasion, and emt of breast cancer cells by directly targeting S100A2. Oncol Res, 2021, 28(7): 731-744. [Crossref]
97. Zhao J, Li C, Qin T, Jin Y, He R, Sun Y, et al. Mechanical overloading-induced miR-325-3p reduction promoted chondrocyte senescence and exacerbated facet joint degeneration. Arthritis Res Ther, 2023, 25(1): 54-67. [Crossref]
98. Lindholm E, Leivonen S, Undlien E, Nebdal D, Git A, Caldas C, et al. miR-342-5p as a potential regulator of HER2 breast cancer cell growth. Microrna, 2019, 8(2): 155-165. [Crossref]
99. Owczarz M, Połosak J, Domaszewska-Szostek A, Kołodziej P, Kuryłowicz A, & Puzianowska-Kuźnicka M. Age-related epigenetic drift deregulates SIRT6 expression and affects its downstream genes in human peripheral blood mononuclear cells. Epigenetics, 2020, 15(12): 1336-1347. [Crossref]
100. Guo D, Ye Y, Qi J, Tan X, Zhang Y, Ma Y, et al. Age and sex differences in microRNAs expression during the process of thymus aging. Acta Biochim Biophys Sin (Shanghai), 2017, 49(5): 409-419. [Crossref]
101. Zheng S, Li M, Miao K, & Xu H. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J Cell Biochem, 2020, 121(3): 2225-2235. [Crossref]
102. Kooistra S, Nørgaard L, Lees M, Steinhauer C, Johansen J, & Helin K. A screen identifies the oncogenic micro-RNA miR-378a-5p as a negative regulator of oncogene-induced senescence. PLoS One, 2014, 9(3): e91034. [Crossref]
103. Zeng X, Ma X, Guo H, Wei L, Zhang Y, Sun C, et al. MicroRNA-582-5p promotes triple-negative breast cancer invasion and metastasis by antagonizing CMTM8. Bioengineered, 2021, 12(2): 10126-10135. [Crossref]
104. Zhang Y, Li X, Cao X, Ji C, Gao X, Gao D, et al. MicroRNA-582-5p contributes to the maintenance of neural stem cells through inhibiting secretory protein FAM19A1. Front Cell Neurosci, 2022, 16: 866020. [Crossref]
105. Gao W, Yuan L, Zhang Y, Huang F, Ai C, Lv T, et al. miR-1246-overexpressing exosomes improve UVB-induced photoaging by activating autophagy via suppressing GSK3β. Photochem Photobiol Sci, 2024, 23(5): 957-972. [Crossref]
106. Zhou B, Xue J, Wu R, Meng H, Li R, Mo Z, et al. CREBZF mRNA nanoparticles suppress breast cancer progression through a positive feedback loop boosted by circPAPD4. J Exp Clin Cancer Res, 2023, 42(1): 138-147. [Crossref]
107. Dell'Aversana C, Cuomo F, Longobardi S, D'Hooghe T, Caprio F, Franci G, et al. Age-related miRNome landscape of cumulus oophorus cells during controlled ovarian stimulation protocols in IVF cycles. Hum Reprod, 2021, 36(5): 1310-1325. [Crossref]
108. Turashvili G, Lightbody E, Tyryshkin K, SenGupta S, Elliott B, Madarnas Y, et al. Novel prognostic and predictive microRNA targets for triple-negative breast cancer. Faseb j, 2018: fj201800120R. [Crossref]
109. Chen Z, Gong X, Cheng C, Fu Y, Wu W, & Luo Z. Circ_0001777 affects triple-negative breast cancer progression through the miR-95-3p/AKAP12 axis. Clin Breast Cancer, 2023, 23(2): 143-154. [Crossref]
110. Yusof K, Groen K, Rosli R, Abdullah M, Mahmud R, & Avery-Kiejda K. Evaluation of circulating micrornas and adipokines in breast cancer survivors with arm lymphedema. Int J Mol Sci, 2022, 23(19): [Crossref]
111. Torrisi R, Vaira V, Giordano L, Fernandes B, Saltalamacchia G, Palumbo R, et al. Identification of a panel of miRNAs associated with resistance to palbociclib and endocrine therapy. Int J Mol Sci, 2024, 25(3): 1498-1506. [Crossref]
112. Chacolla-Huaringa R, Moreno-Cuevas J, Trevino V, & Scott S. Entrainment of breast cell lines results in rhythmic fluctuations of microRNAs. Int J Mol Sci, 2017, 18(7): 1499-1507. [Crossref]
113. Yuan C. miR-616 promotes breast cancer migration and invasion by targeting TIMP2 and regulating MMP signaling. Oncol Lett, 2019, 18(3): 2348-2355. [Crossref]
114. Wang B, Wang Y, Wang X, Gu J, Wu W, Wu H, et al. Extracellular vesicles carrying miR-887-3p promote breast cancer cell drug resistance by targeting BTBD7 and activating the Notch1/Hes1 signaling pathway. Dis Markers, 2022, 2022: 5762686. [Crossref]
115. Abd E, Ghanem H, Swellam M, & Taha A. Involvement of FAM170B-AS1, hsa-miR-1202, and hsa-miR-146a-5p in breast cancer. Cancer Biomark, 2024, 39(4): 313-333. [Crossref]
116. Klinge C, Piell K, Tooley C, & Rouchka E. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep, 2019, 9(1): 9430-9445. [Crossref]
117. Lv Z, Yang K, & Wang Y. Long non-coding RNA breast cancer-associated transcript 54 sponges microRNA-1269b to suppress the proliferation of hemangioma-derived endothelial cells. Bioengineered, 2022, 13(3): 6188-6195. [Crossref]
118. Peng Z, Xu B, & Jin F. Circular RNA hsa_circ_0000376 participates in tumorigenesis of breast cancer by targeting miR-1285-3p. Technol Cancer Res Treat, 2020, 19: 1533033820928471. [Crossref]
119. Zhao D, Wu K, Sharma S, Xing F, Wu S, Tyagi A, et al. Exosomal miR-1304-3p promotes breast cancer progression in African Americans by activating cancer-associated adipocytes. Nat Commun, 2022, 13(1): 7734-7745. [Crossref]
120. Tu D, Dou J, Wang M, Zhuang H, & Zhang X. M2 macrophages contribute to cell proliferation and migration of breast cancer. Cell Biol Int, 2021, 45(4): 831-838. [Crossref]
121. He Y, Xiao B, Qiu J, Lei T, Li L, & Sun Z. Study on the expression of microRNA-1825 in serum of pre-operative and post-operative patients with breast cancer. Zhonghua Yu Fang Yi Xue Za Zhi, 2021, 55(5): 691-697. [Crossref]
122. Singh J, Sangwan N, Chauhan A, & Avti P. Integrative expression, survival analysis and cellular miR-2909 molecular interplay in MRN complex check point sensor genes (MRN-CSG) involved in breast cancer. Clin Breast Cancer, 2022, 22(8): e850-e862. [Crossref]
123. Dong G, Wang X, Jia Y, Jia Y, Zhao W, Zhang J, et al. HAND2-AS1 works as a ceRNA of miR-3118 to suppress proliferation and migration in breast cancer by upregulating PHLPP2. Biomed Res Int, 2020, 2020: 8124570. [Crossref]
124. Al-Khanbashi M, Caramuta S, Alajmi A, Al-Haddabi I, Al-Riyami M, Lui W, et al. Tissue and serum miRNA profile in locally advanced breast cancer (LABC) in response to neo-adjuvant chemotherapy (NAC) treatment. PLoS One, 2016, 11(4): e0152032. [Crossref]
125. Lou W, Liu J, Ding B, Xu L, & Fan W. Identification of chemoresistance-associated miRNAs in breast cancer. Cancer Manag Res, 2018, 10: 4747-4757. [Crossref]
126. Li Y, Shan F, & Chen J. Lipid raft-mediated miR-3908 inhibition of migration of breast cancer cell line MCF-7 by regulating the interactions between AdipoR1 and Flotillin-1. World J Surg Oncol, 2017, 15(1): 69-80. [Crossref]
127. Carvalho T, Brasil G, Jucoski T, Adamoski D, de Lima R, Spautz C, et al. MicroRNAs miR-142-5p, miR-150-5p, miR-320a-3p, and miR-4433b-5p in serum and tissue: potential biomarkers in sporadic breast cancer. Front Genet, 2022, 13: 865472. [Crossref]
128. Boo L, Ho W, Ali N, Yeap S, Ky H, Chan K, et al. MiRNA transcriptome profiling of spheroid-enriched cells with cancer stem cell properties in human breast MCF-7 cell line. Int J Biol Sci, 2016, 12(4): 427-445. [Crossref]
129. Kwak S, Yoo J, An H, Bae I, Park M, Kim J, et al. miR-5003-3p promotes epithelial-mesenchymal transition in breast cancer cells through Snail stabilization and direct targeting of E-cadherin. J Mol Cell Biol, 2016, 8(5): 372-383. [Crossref]
130. Tian X, Yu H, Li D, Jin G, Dai S, Gong P, et al. The miR-5694/AF9/snail axis provides metastatic advantages and a therapeutic target in basal-like breast cancer. Mol Ther, 2021, 29(3): 1239-1257. [Crossref]
131. Satomi-Tsushita N, Shimomura A, Matsuzaki J, Yamamoto Y, Kawauchi J, Takizawa S, et al. Serum microRNA-based prediction of responsiveness to eribulin in metastatic breast cancer. PLoS One, 2019, 14(9): e0222024. [Crossref]
132. Gao Y, Ma H, Gao C, Lv Y, Chen X, Xu R, et al. Tumor-promoting properties of miR-8084 in breast cancer through enhancing proliferation, suppressing apoptosis and inducing epithelial-mesenchymal transition. J Transl Med, 2018, 16(1): 38-50. [Crossref]
133. Esposito M, Gualandi N, Spirito G, Ansaloni F, Gustincich S, & Sanges R. Transposons acting as competitive endogenous rnas: In-silico evidence from datasets characterised by L1 overexpression. Biomedicines, 2022, 10(12): 3279-3289. [Crossref]
134. Moro J, Grinpelc A, Farré P, Duca R, Lacunza E, Graña K, et al. miR-877-5p as a potential link between triple-negative breast cancer development and metabolic syndrome. Int J Mol Sci, 2023, 24(23): 16758. [Crossref]
135. Ding J, Wu W, Yang J, & Wu M. Long non-coding RNA MIF-AS1 promotes breast cancer cell proliferation, migration and EMT process through regulating miR-1249-3p/HOXB8 axis. Pathol Res Pract, 2019, 215(7): 152376. [Crossref]
136. Ma L, Zhang Y, & Hu F. miR‑28‑5p inhibits the migration of breast cancer by regulating WSB2. Int J Mol Med, 2020, 46(4): 1562-1570. [Crossref]
137. Chen Q, Xu H, Zhu J, Feng K, & Hu C. LncRNA MCM3AP-AS1 promotes breast cancer progression via modulating miR-28-5p/CENPF axis. Biomed Pharmacother, 2020, 128: 110289. [Crossref]
138. Zan X, Li W, Wang G, Yuan J, Ai Y, Huang J, et al. Circ-CSNK1G1 promotes cell proliferation, migration, invasion and glycolysis metabolism during triple-negative breast cancer progression by modulating the miR-28-5p/LDHA pathway. Reprod Biol Endocrinol, 2022, 20(1): 138-148. [Crossref]
139. Poodineh J, Sirati-Sabet M, Rajabibazl M, & Mohammadi-Yeganeh S. MiR-130a-3p blocks Wnt signaling cascade in the triple-negative breast cancer by targeting the key players at multiple points. Heliyon, 2020, 6(11): e05434. [Crossref]
140. Xie D, Li S, Wu T, Wang X, & Fang L. MiR-181c suppresses triple-negative breast cancer tumorigenesis by targeting MAP4K4. Pathol Res Pract, 2022, 230: 153763. [Crossref]
141. Wang M, Liu Z, Zhang H, Liu H, & Ma L. Hsa_circRNA_000166 accelerates breast cancer progression via the regulation of the miR-326/ELK1 and miR-330-5p/ELK1 axes. Ann Med, 2024, 56(1): 2424515. [Crossref]
142. Yu S, Zhou Y, Niu L, Qiao Y, & Yan Y. Mesenchymal stem cell-derived exosome mir-342-3p inhibits metastasis and chemo-resistance of breast cancer through regulating ID4. Genes Genomics, 2022, 44(5): 539-550. [Crossref]
143. Zhou D, Ren K, Wang M, Wang J, Li E, Hou C, et al. Long non-coding RNA RACGAP1P promotes breast cancer invasion and metastasis via miR-345-5p/RACGAP1-mediated mitochondrial fission. Mol Oncol, 2021, 15(2): 543-559. [Crossref]
144. Hao S, Tian W, Chen Y, Wang L, Jiang Y, Gao B, et al. MicroRNA-374c-5p inhibits the development of breast cancer through TATA-box binding protein associated factor 7-mediated transcriptional regulation of DEP domain containing 1. J Cell Biochem, 2019, 120(9): 15360-15368. [Crossref]
145. Zou Y, Chen Y, Yao S, Deng G, Liu D, Yuan X, et al. MiR-422a weakened breast cancer stem cells properties by targeting PLP2. Cancer Biol Ther, 2018, 19(5): 436-444. [Crossref]
146. Zhao L, Feng X, Song X, Zhou H, Zhao Y, Cheng L, et al. miR-493-5p attenuates the invasiveness and tumorigenicity in human breast cancer by targeting FUT4. Oncol Rep, 2016, 36(2): 1007-1015. [Crossref]
147. Qiu X, Zhang Q, Deng Q, & Li Q. Circular RNA hsa_circ_0012673 promotes breast cancer progression via miR-576-3p/SOX4 Axis. Mol Biotechnol, 2023, 65(1): 61-71. [Crossref]
148. Sun L, Chen S, Wang T, & Bi S. Hsa_circ_0008673 promotes breast cancer progression by MiR-578/GINS4 axis. Clin Breast Cancer, 2023, 23(3): 281-290. [Crossref]
149. Yuan D, Liu J, Sang W, & Li Q. Comprehensive analysis of the role of SFXN family in breast cancer. Open Med, 2023, 18(1): 20230685. [Crossref]
150. Zhang Z, Luo X, Xue X, Pang M, Wang X, Yu L, et al. Engineered exosomes carrying miR-588 for treatment of triple negative breast cancer through remodeling the immunosuppressive microenvironment. Int J Nanomedicine, 2024, 19: 743-758. [Crossref]
151. Choi S, An H, Yeo H, Sung M, Oh J, Lee K, et al. MicroRNA‑606 inhibits the growth and metastasis of triple‑negative breast cancer by targeting Stanniocalcin 1. Oncol Rep, 2024, 51(1): 8661. [Crossref]
152. Yang T, Yu R, Cheng C, Huo J, Gong Z, Cao H, et al. Cantharidin induces apoptosis of human triple negative breast cancer cells through mir-607-mediated downregulation of EGFR. J Transl Med, 2023, 21(1): 597-606. [Crossref]
153. Bhajun R, Guyon L, Pitaval A, Sulpice E, Combe S, Obeid P, et al. A statistically inferred microRNA network identifies breast cancer target miR-940 as an actin cytoskeleton regulator. Sci Rep, 2015, 5: 8336-8346. [Crossref]
154. Shi G, Li H, Chen Y, Chen Z, & Lin X. CircSEPT9 promotes breast cancer progression by regulating PTBP3 expression via sponging miR-625-5p. Thorac Cancer, 2024, 15(10): 808-819. [Crossref]
155. Ren F, Rui X, & Xiao X. Loss of miR-634 contributes to the formation FOXA1-positive triple negative breast cancer subtype. Discov Oncol, 2024, 15(1): 584-595. [Crossref]
156. Tang C, Wang X, Ji C, Zheng W, Yu Y, Deng X, et al. The role of miR-640: a potential suppressor in breast cancer via Wnt7b/β-catenin signaling pathway. Front Oncol, 2021, 11: 645682. [Crossref]
157. Darvishi N, Rahimi K, Mansouri K, Fathi F, Menbari M, Mohammadi G, et al. MiR-646 prevents proliferation and progression of human breast cancer cell lines by suppressing HDAC2 expression. Mol Cell Probes, 2020, 53: 101649. [Crossref]
158. Lee J, Guan W, Han S, Hong D, Kim L, & Kim H. MicroRNA-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Science, 2018, 109(5): 1404-1413. [Crossref]
159. Dong H, Liu Q, Zhao T, Yao F, Xu Y, Chen B, et al. Long non-coding RNA LOXL1-AS1 drives breast cancer invasion and metastasis by antagonizing miR-708-5p expression and activity. Mol Ther Nucleic Acids, 2020, 19: 696-705. [Crossref]
160. Zhou X, Jian W, Luo Q, Zheng W, Deng X, Wang X, et al. Circular RNA_0006014 promotes breast cancer progression through sponging miR-885-3p to regulate NTRK2 and PIK3/AKT pathway. Aging, 2022, 14(7): 3105-3128. [Crossref]
161. Xu C, Yu H, Yin X, Zhang J, Liu C, Qi H, et al. Circular RNA circNINL promotes breast cancer progression through activating β-catenin signaling via miR-921/ADAM9 axis. J Biochem, 2021, 169(6): 693-700. [Crossref]
162. Ji C, Zhu L, & Fang L. Hsa_circ_0000851 promotes PDK1/p-AKT-mediated cell proliferation and migration by regulating miR-1183 in triple-negative breast cancer. Cell Signal, 2023, 101: 110494. [Crossref]
163. Torkashvand S, Damavandi Z, Mirzaei B, Tavallaei M, Vasei M, & Mowla S. Decreased expression of bioinformatically predicted piwil2-targetting microRNAs, miR-1267 and miR-2276 in breast cancer. Arch Iran Med, 2016, 19(6): 420-425.
164. Sameer C, Shah M, Nandy D, & Gupta R. Genomic index of sensitivity to chemotherapy for triple negative breast cancer. Asian Pac J Cancer Prev, 2023, 24(6): 2043-2053. [Crossref]
165. Du H, & Liu B. MiR-1271 as a tumor suppressor in breast cancer proliferation and progression via targeting SPIN1. Eur Rev Med Pharmacol Sci, 2018, 22(9): 2697-2706. [Crossref]
166. Yu T, Yu H, Sun J, Zhao Z, Li S, Zhang X, et al. miR-1271 inhibits ERα expression and confers letrozole resistance in breast cancer. Oncotarget, 2017, 8(63): 107134-107148. [Crossref]
167. Hironaka-Mitsuhashi A, Otsuka K, Gailhouste L, Sanchez Calle A, Kumazaki M, Yamamoto Y, et al. MiR-1285-5p/TMEM194A axis affects cell proliferation in breast cancer. Cancer Sci, 2020, 111(2): 395-405. [Crossref]
168. Chen K, Xiao X, & Xu Z. MiR-1294 inhibits the progression of breast cancer via regulating ERK signaling. Bull Cancer, 2022, 109(10): 999-1006. [Crossref]
169. Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer, 2020, 19(1): 85-96. [Crossref]
170. Yu C, Zhang T, Chen F, & Yu Z. The impact of hsa-miR-1972 on the expression of von Willebrand factor in breast cancer progression regulation. PeerJ, 2024, 12: e18476. [Crossref]
171. Yu L, Zhang W, Wang P, Zhang Q, Cong A, Yang X, et al. LncRNA SNHG11 aggravates cell proliferation and migration in triple-negative breast cancer via sponging miR-2355-5p and targeting CBX5. Exp Ther Med, 2021, 22(2): 892-904. [Crossref]
172. Luo X, & Wang H. LINC00514 upregulates CCDC71L to promote cell proliferation, migration and invasion in triple-negative breast cancer by sponging miR-6504-5p and miR-3139. Cancer Cell Int, 2021, 21(1): 180-191. [Crossref]
173. Delgir S, Ilkhani K, Safi A, Rahmati Y, Montazari V, Zaynali-Khasraghi Z, et al. The expression of miR-513c and miR-3163 was downregulated in tumor tissues compared with normal adjacent tissue of patients with breast cancer.BMC Med Genomics, 2021, 14(1): 180-190. [Crossref]
174. Wei M, Yu H, Cai C, Gao R, Liu X, & Zhu H. MiR-3194-3p inhibits breast cancer progression by targeting Aquaporin1. Front Oncol, 2020, 10: 1513-1522. [Crossref]
175. Akshaya R, Akshaya N, & Selvamurugan N. A computational study of non-coding RNAs on the regulation of activating transcription factor 3 in human breast cancer cells. Comput Biol Chem, 2020, 89: 107386. [Crossref]
176. Wang B, Li J, Sun M, Sun L, & Zhang X. miRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features. IUBMB Life, 2014, 66(5): 371-377. [Crossref]
177. Li Y, Wang Y, Chen X, Ma R, Guo X, Liu H, et al. MicroRNA-4472 promotes tumor proliferation and aggressiveness in breast cancer by targeting RGMA and inducing EMT. Clin Breast Cancer, 2020, 20(2): e113-e126. [Crossref]
178. Shin D, Yoo J, Jeong J, & Han Y. MiR-5586-5p suppresses hypoxia-induced angiogenesis through multiple targeting of HIF-1α, HBEGF and ADAM17 in breast cancer. Anticancer Res, 2025, 45(2): 473-489. [Crossref]
179. Wang D, Yang S, Lyu M, Xu L, Zhong S, & Yu D. Circular RNA HSDL2 promotes breast cancer progression via miR-7978 ZNF704 axis and regulating hippo signaling pathway. Breast Cancer Res, 2024, 26(1): 105-116. [Crossref]
180. Yentrapalli R, Azimzadeh O, Kraemer A, Malinowsky K, Sarioglu H, Becker K, et al. Quantitative and integrated proteome and microRNA analysis of endothelial replicative senescence. J Proteomics, 2015, 126: 12-23. [Crossref]
181. Yoo J, Kim C, Jung H, Lee D, & Kim J. Discovery and characterization of miRNA during cellular senescence in bone marrow-derived human mesenchymal stem cells. Exp Gerontol, 2014, 58: 139-145. [Crossref]
182. Dalton S, Smith K, Singh K, Kaiser H, Kolhe R, Mondal A, et al. Accumulation of kynurenine elevates oxidative stress and alters microRNA profile in human bone marrow stromal cells./em> Exp Gerontol, 2020, 130: 110800. [Crossref]
183. Chen J, Zou Q, Lv D, Wei Y, Raza M, Chen Y, et al. Comprehensive transcriptional landscape of porcine cardiac and skeletal muscles reveals differences of aging. Oncotarget, 2018, 9(2): 1524-1541. [Crossref]
184. Mennitti L, Carpenter A, Loche E, Pantaleão L, Fernandez-Twinn D, Schoonejans J, et al. Effects of maternal diet-induced obesity on metabolic disorders and age-associated miRNA expression in the liver of male mouse offspring. Int J Obes (Lond), 2022, 46(2): 269-278. [Crossref]
185. Guo Y, Tian L, Liu X, He Y, Chang S, & Shen Y. ERRFI1 inhibits proliferation and inflammation of nucleus pulposus and is negatively regulated by miR-2355-5p in intervertebral disc degeneration. Spine (Phila Pa 1976), 2019, 44(15): E873-e881. [Crossref]
186. Morsiani C, Bacalini M, Collura S, Moreno-Villanueva M, Breusing N, Bürkle A, et al. Blood circulating miR-28-5p and let-7d-5p associate with premature ageing in down syndrome. Mech Ageing Dev, 2022, 206: 111691. [Crossref]
187. Manzano-Crespo M, Atienza M, & Cantero JL. Lower serum expression of miR-181c-5p is associated with increased plasma levels of amyloid-beta 1-40 and cerebral vulnerability in normal aging. Transl Neurodegener, 2019, 8: 34-46. [Crossref]
188. Schneider A, Matkovich S, Victoria B, Spinel L, Bartke A, Golusinski P, et al. Changes of ovarian microRNA profile in long-living ames dwarf mice during aging. PLoS One, 2017, 12(1): e0169213. [Crossref]
189. Wang L, Si X, Chen S, Wang X, Yang D, Yang H, et al. A comprehensive evaluation of skin aging-related circular RNA expression profiles. J Clin Lab Anal, 2021, 35(4): e23714. [Crossref]
190. Zhang J, Gong H, Zhao T, Xu W, Chen H, Li T, et al. AMPK-upregulated microRNA-708 plays as a suppressor of cellular senescence and aging via downregulating disabled-2 and mTORC1 activation. MedComm, 2024, 5(3): e475. [Crossref]
191. Yang Y, Zhang W, Wang X, Yang J, Cui Y, Song H, et al. A passage-dependent network for estimating the in vitro senescence of mesenchymal stromal/stem cells using microarray, bulk and single cell RNA sequencing. Front Cell Dev Biol, 2023, 11: 998666. [Crossref]
192. Huang W, Lin Q, Chen S, Sun L, Chen Q, Yi K, et al. Correction for: Integrated analysis of microRNA and mRNA expression profiling identifies BAIAP3 as a novel target of dysregulated hsa-miR-1972 in age-related white matter lesions. Aging, 2021, 13(11): 15688-15689. [Crossref]