Open Access | Research
This work is licensed under a
Creative Commons Attribution-ShareAlike 4.0 International License.
Molecular docking and ADMET analysis of coenzyme Q10 as a potential therapeutic agent for Alzheimer's disease
* Corresponding author: Abdullah Al Noman
Mailing address: School of Pharmacy, BRAC University, Dhaka, Bangladesh.
Email: abdullah.al.noman@g.bracu.ac.bd
This article belongs to the Special Issue:
Nutrient compounds for intervention of aging and age-related diseases
Received: 09 October 2024 / Revised: 29 October 2024 / Accepted: 30 October 2024 / Published: 28 December 2024
DOI: 10.31491/APT.2024.12.155
Abstract
Alzheimer's disease (AD) is a neurodegenerative disorder characterized by cognitive decline, synaptic dysfunction, and neuroinflammation, with oxidative stress playing a crucial role. Coenzyme Q10 (CoQ10), known for its antioxidant properties, has been proposed as a potential therapeutic agent due to its ability to mitigate oxidative damage and maintain mitochondrial integrity. This study investigates the molecular interactions of CoQ10 with key proteins involved in AD pathology—glycogen synthase kinase-3β (GSK-3β), protein kinase B (PKB), and phosphoinositide 3-kinase (PI3K)—through molecular docking methods. Results indicate that CoQ10 exhibits the strongest binding affinity with GSK-3β, potentially reducing tau protein phosphorylation, a hallmark of AD. ADMET analysis further supports CoQ10's drug-like properties, with a favorable absorption and safety profile, although limited blood-brain barrier permeability poses a challenge. This in silico study highlights CoQ10's therapeutic potential in addressing neuroinflammation and oxidative stress in AD, warranting further in vitro and in vivo validation.
Keywords
Alzheimer's disease, coenzyme Q10, molecular docking, oxidative stress, neuroinflammation
Introduction
Alzheimer's disease (AD) is a chronic, progressive neurodegenerative condition predominately affecting the elderly, which causes substantial
deficits in memory, cognitive deterioration, and synaptic connections difficulties [1]. It represents
60-80% of global dementia cases, and despite numerous research initiatives, no specific cure has been uncovered [2]. The condition known as Alzheimer's pathogenesis is characterized by
two primary features: exogenous amyloid-beta (Aβ) plaques and intracellular neurofibrillary knots that consist of hyperphosphorylated
tau proteins. For these characteristics, two essential pathogenic mechanisms—neuroinflammation and oxidative stress—are important to the
disease's development [3].
Neuroinflammation is the continuous activation of microglia and astrocytes, which produce pro-inflammatory cytokines that exacerbate
damage to the neurons [4].
Oxidative stress is caused by an imbalance between the formation of
reactive oxygen species (ROS) and the brain's defense against antioxidants, which causes damage to lipids, proteins, and genetic
material [5].
Mitochondrial dysfunction has grown into an important contributor to oxidative stress in the development of AD [6]. Mitochondria, as the essential suppliers of cellular energy and ROS,
contribute to increased damage from oxidation and neuroinflammation when
malfunctioning. Coenzyme Q10 (CoQ10) is a possible chemical substance in
the fight against oxidative stress, functioning as a naturally occurring
antioxidant and a crucial component of the chain of electron transport in
the mitochondria. CoQ10 works both in cellular production of energy and in
neutralizing free radicals, therefore preserving neurons from damage
caused by oxidation and protecting mitochondrial integrity. Additionally,
CoQ10 has demonstrated neuroprotective properties in numerous
neurodegenerative conditions, which include Parkinson's and Huntington's
diseases, by lowering oxidative damage and inhibiting caspase pathways [7]. The neuroprotective impact of CoQ10 in the progression of Alzheimer's
has been found to be connected to the ability to regulate important
signaling pathways related to surviving cells and inflammation. The PI3K/Akt pathway plays an important role in improving neuronal life
expectancy through the regulation of apoptosis, autophagy, respectively, and the inflammatory response. Three essential proteins in this
pathway—glycogen synthase kinase-3 beta (the transcription factor GSK),
protein kinase B, also known as PKB/Akt, and phosphoinositide 3-kinase (PI3K)—are particularly noteworthy in the study of Alzheimer's research.
GSK-3β contributes to the degradation of tau proteins, which produces neurofibrillary tangles, while PI3K and Akt perform protective roles by
enhancing cell survival and preventing apoptosis. The process of deregulation of this pathway has been associated with the development of
AD and increased susceptibility to damage from oxidation and inflammation [8]. Using a molecular docking
method, this research is aimed at investigating the potential of CoQ10 to
mitigate oxidative stress and neuroinflammation in AD, with particular emphasis on its connections with GSK-3β, PKB/Akt, and PI3K. An effective
way to investigate possible relationships between CoQ10 and target proteins, as well as potential regulation of the PI3K/Akt pathway, is by
using the computational methodology of molecular docking. By analyzing CoQ10's impact on the proteins that signal associated with the major
pathogenic mechanisms of AD, we expect to contribute molecular insights into the drug's potential. By comprehension of these molecular
interactions, CoQ10 may become an alternative therapeutic intervention candidate in AD, providing a new approach to control neuroinflammation and
oxidative stress. Beside that in the animal model, CoQ10 has been shown to have evidence of decreasing amyloid beta levels and formation of plaque.
In animals, this explains why it may play a prominent role in mitigating AD pathology [9,
10].
Materials and methods
Molecular docking study
Computer-based tools were used to visualize the protein and ligand capabilities. These tools are very effective in molecular docking and save time [11]. Various online tools and software were used for this study, such as PyRx 0.8, ChimeraX 1.8, BIOVIA Discovery Studio 2024 Client, PkCSM, Errat, and Pro-SA Web.
Ligand selection
For this study, CoQ10 was selected. CoQ10 is an influential antioxidant that is produced by our body inherently [12]. This coenzyme has significant antioxidant properties. This coenzyme can be used as an important bioactive compound to relieve the oxidation [13]. The antioxidant character of CoQ10 helps to remove oxidative stress from cells. Oxidative stress is mainly connected to AD [14, 15]. Moreover, some research has found that CoQ10 plays a significant role in decreasing amyloid-β plaques and tau protein tangles; these factors are connected to AD [15, 16]. Overall, this compound was thought to be perfect for the study.
Protein selection
CoQ10 interacts with a number of important proteins involved in IL-17-mediated neurological inflammation and oxidative stress in AD [17, 18]. The selected proteins are 1. phosphoinositide 3-kinase (PI3K) (PDB ID: 3APD), 2. protein kinase B (PKB) (PDB ID: 1UNQ), and 3. glycogen synthase kinase-3β (GSK-3β) (PDB ID: 1GNG). PI3K and PKB are parts of the PI3K and Akt signaling pathways. These proteins have a significant impact on neuroprotection and survival of cells [19]. Further, GSK-3β is involved in tau protein phosphorylation [19].
Ligand preparation
From previous findings, CoQ10 (PDB ID: 521915) was selected [20]. The structure of this compound was collected from Pubchem (https://pubchem.ncbi.nlm.nih.gov/, accessed on October 4, 2024) in SDF format for binding to selected proteins. To start with, the coenzyme was directly inserted in the PyRx software. Other software recommends the pdbqt format, but in PyRx, there is no need for the pdbqt file, because PyRx directly supports sdf files [21]. After that, energy was reduced of that compound for a perfect result. For docking, the compound was then converted into pdbqt format. Science PyRx has access to openbable [22]. After performing docking, the specific conformation of this compound is selected and saved as a PDB file for further analysis.
Protein check
Errat
The accuracy and reliability of the proteins were detected by ERRAT (https://saves.mbi.ucla.edu/, accessed on October 4, 2024), an online program that analyzes the quality of a protein structure by examining non-bonded atomatom interactions [23]. An overall quality factor greater than 90% shows a high quality model [24]. According to Errat, the overall quality factor of PI3K (PDB ID: 3APD) was 95.641% (Figure 1C), PKB (PDB ID: 1UNQ) was 95.327% (Figure 1B), and GSK-3β (PDB ID: 1GNG) was 92.222% (Figure 1A). The overall quality factors of all proteins are above 90%, which means high-quality model. Based on Figure 1, the x-axis represents all residues and the y-axis represents the errors. The red lines show the higher error value, which is above 99% or only at 99%, and the yellow vertical lines also indicate the error value, but at a low level. Moreover, the two horizontal lines at 95% and 99% are the threshold lines.
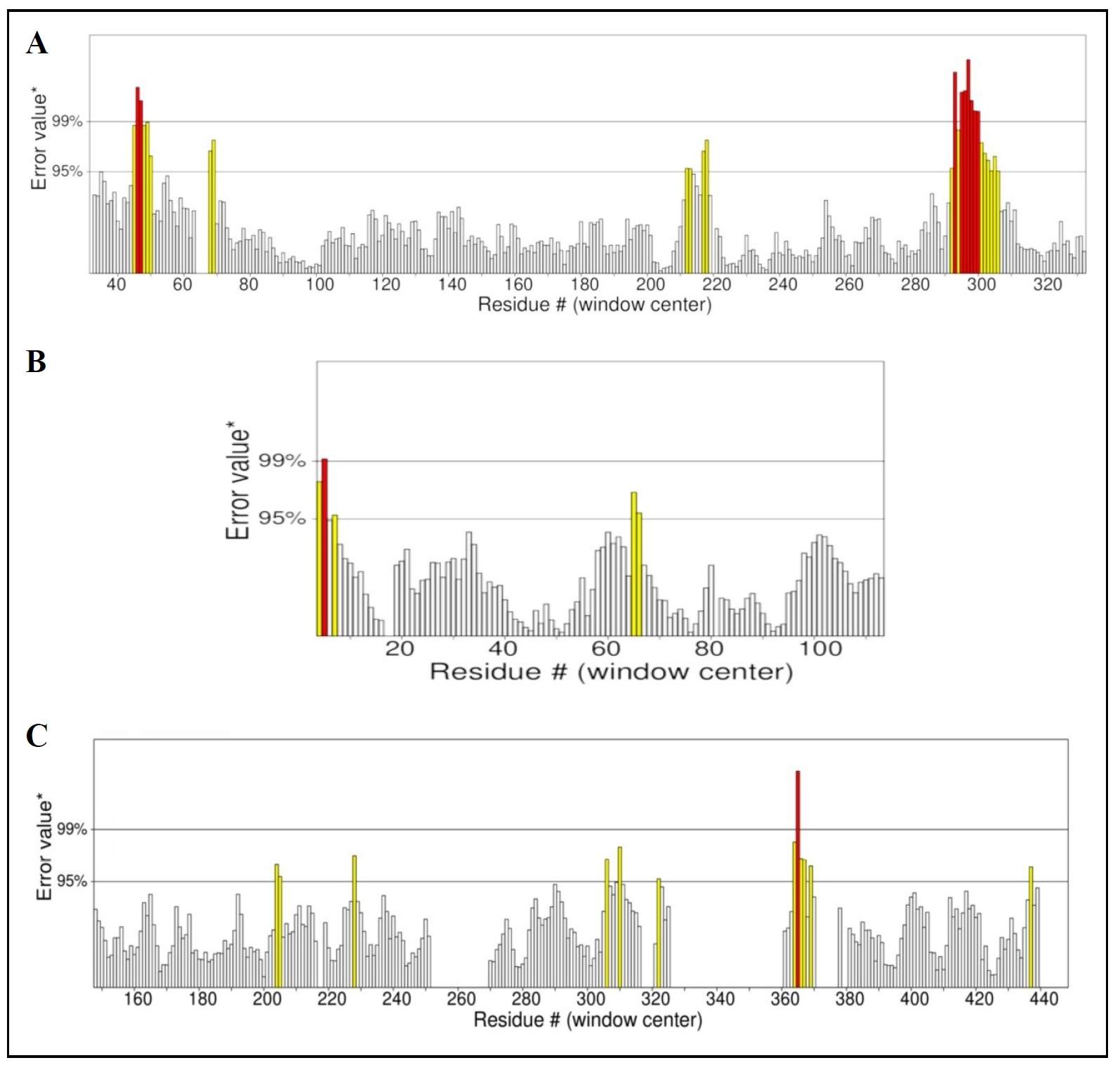
Figure 1. Error values distribution over all residues. The overall quality factor of GSK-3β (A), PKB (B), and PI3K (C).
ProSA-Web
Moreover, the ProSa-Web tool (https://prosa.services.came.sbg.ac.at/prosa.php, accessed on October 4, 2024) was used to check the proteins. It is a
useful online tool that finds problems in three-dimensional protein
structures. It also gives score and energy graphs to highlight faults in
protein models and the results are shown in graphs (Figure 2
and Figure 3) [25]. The more negative value of the z-score represents a better protein
model [26,27].
According to Figure 2, the
z-score for GSK-3β is -8.68, which indicated that the protein is located
in the NMR solved protein structure (Figure 2A); PKB is -4.97, which represented that the protein is present in the NMR
solved protein structure (Figure 2B); and PI3K is -12.1, which showed that the protein structure is
situating in X-ray solved protein structure (Figure 2C). The z-scores are represented as black dots in the graphs. Besides, the
x-axis and y-axis of the energy plots (Figure 3) show the position of the amino acid sequences and the energy values. On
the y-axis, positive values show the problematic regions and negative
values represent the correct regions of a protein. In Figure 3, it is
noticeable that GSK-3beta (Figure 3A) and PKB (Figure 3B) have more
negative energy levels than PI3K (Figure 3C).
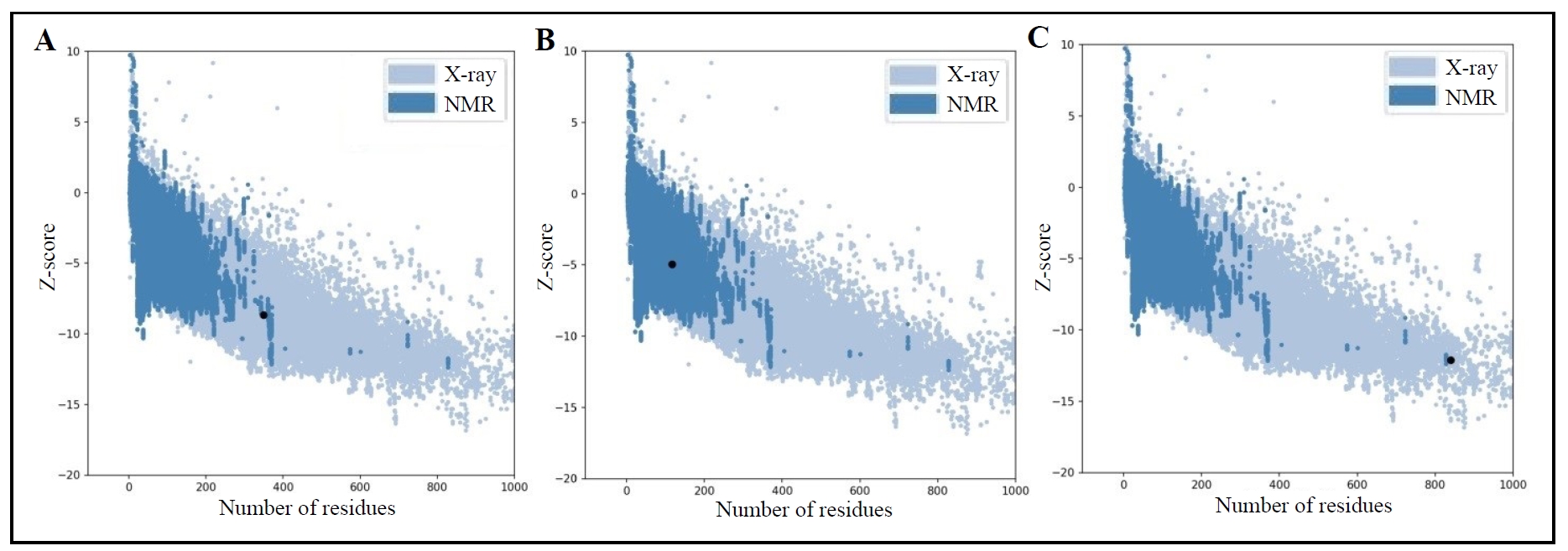
Figure 2. Z-score graphs for identified proteins, including GSK-3β (A), PKB (B), and PI3K (C).
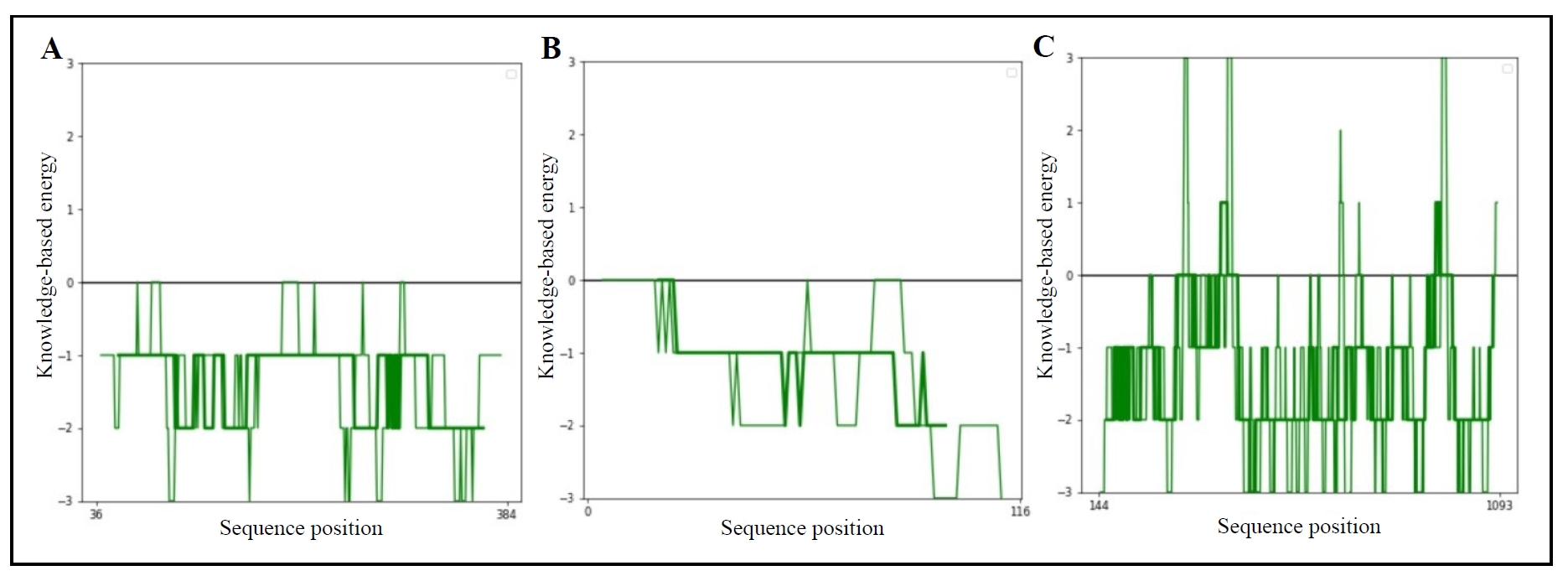
Figure 3. Z-score graphs for identified proteins, including GSK-3β (A), PKB (B), and PI3K (C).
Hydrophobicity
The hydrophobicity graphs in Figure 4 show the hydrophobicity properties of amino acid residues in a protein sequence. The x-axis indicates the position of the amino acid residues and the y-axis represents the average hydrophobicity of each amino acid. Again, the yellow circles exhibit the hydrophobicity of each residue and the blue lines connect the yellow circles to help visualize the data point's position. The positive values above 0 display the hydrophobic areas and are found in the interior of the protein as well as in the transmembrane region. On the other hand, the negative values below 0 show the hydrophilic areas that are present in the interaction with other molecules.
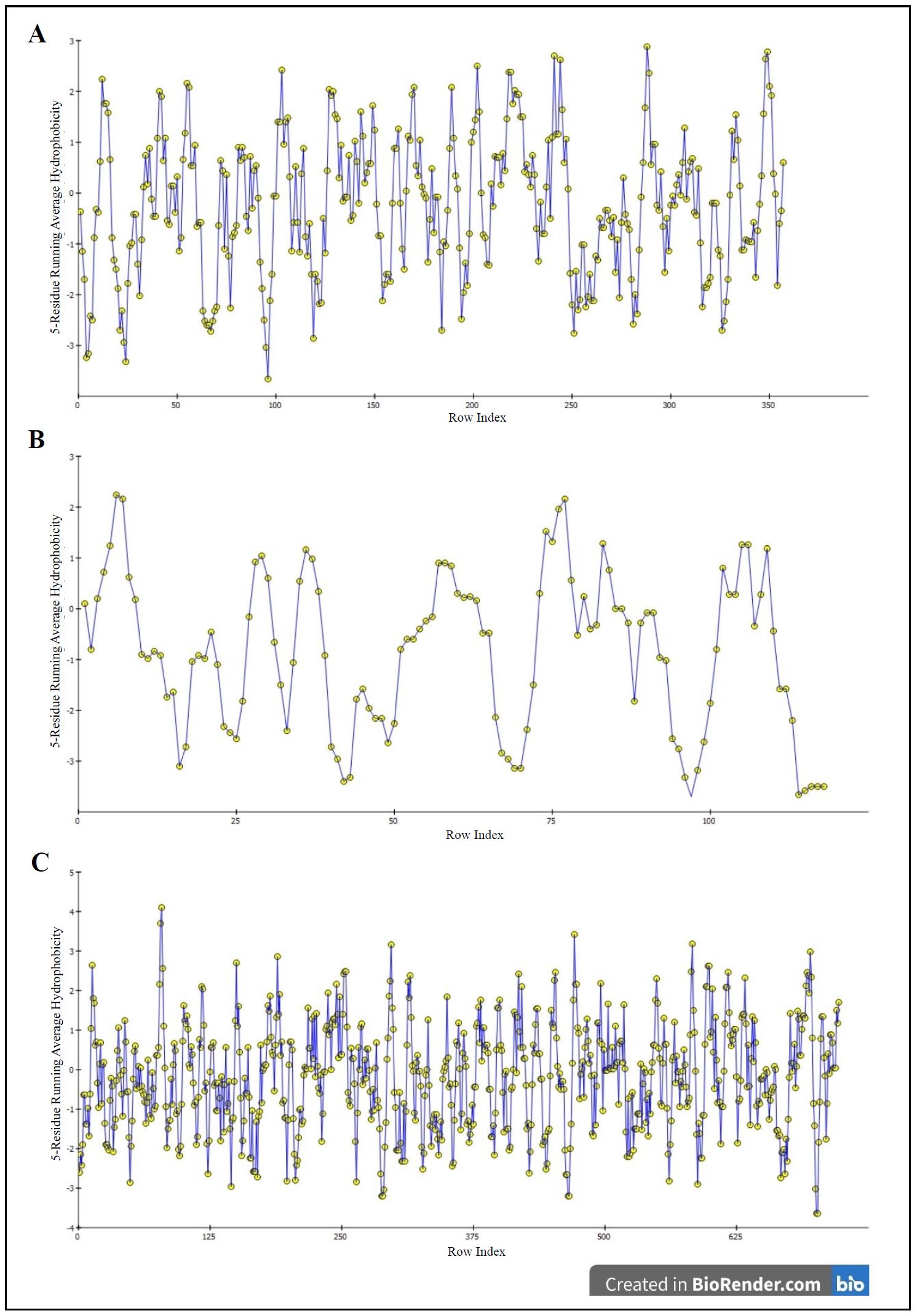
Figure 4. Hydrophobicity graphs for GSK-3β (A), PKB (B), and PI3K (C).
Protein preparation
The identified proteins are prepared one by one using the UCSF ChimeraX software. It is a reliable software for preparing a protein for molecular docking studies [28]. First, a protein's pdb file is inserted in the software. Next, the heteroatoms are deleted, and one chain is selected for the protein, which has two or more protein chains. After that, the Dock Prep option is selected for full preparation. In this section, H-bonds, charges and missing residues are added to the protein [29]. After completing these steps, the newly prepared protein is saved as a pdb file. Furthermore, the active sites of the proteins are identified using Discovery Studio [30].
Protein-ligand binding
Before binding proteins to the specific compounds, their active sites are determined using Discovery Studio. This software is very useful to quickly identify the active sites [31]. For GSK-3β (PDB ID: 1GNG), ARG96, ARG180, LYS205, ARG209, ASN213, VAL214, and VAL208 are the amino acids as active sites. Again, amino acids including SER0, MET1, SER2, HIS13, LYS14, GLY16, GLU17, TYR18, ILE19, ARG23, ARG25, LEU52, ASN53, PHE55, and ARG86 are selected as active binding sites for PKB (PDB ID: 1UNQ). TRP212, LYS288, ILE831, LYS833, ASP836, ILE879, GLU880, ILE881, VAL882, THR887, ASP946, ARG947, HIS948, ASN951, MET953, ILE963, ASP964, and ILE968 are identified for PI3K (PDB ID: 3APD). During molecular docking, ligands do not always bind to the identified active sites of a protein. This situation is known allosteric regulation [32]. For perfect binding, the protein grid box was maximized. As a result, whole proteins become selected and ligands can bind to the perfect site. First, a specific protein and ligand were inserted into PyRx for molecular docking. Next, different positions and dimensions were set for the grid box for maximization due to each protein. The grid box was fixed into center_ x = 31.1276, center_ y = -0.0207, center_ z = 26.3320 and dimension_ x = 81.9806, dimension_ y = -63.7440, dimension_ z = 59.5978 for 3APD. For 1GNG, the grid box was set into center_ x = 108.3264, center_ y = 55.8664, center_ z = 35.0965 and dimension_ x = 36.7377, dimension_ y = 30.3748, dimension_ z = 34.9006. And the box was selected into center_ x = 21.7185, center_ y = 14.4762, center_ z = 9.7653 and dimension_ x = 40.8121, dimension_ y = 38.0033, dimension_ z = 49.1876 for 1UNQ. PyRx has accessibility to Auto-dock Vina to perform docking [33]. After the docking is finished, the significant conformation for the selected compound was saved as a pdb file for each proteinligand binding. During selection, the binding energy for each conformation was determined, and the conformation of a ligand with the lowest binding energy was selected [34]. Finally, the proteins and ligands were entered into the Discovery Studio tool for further analysis [35].
ADMET analysis
To understand the pharmacokinetics, such as absorption, distribution, metabolism, excretion, and toxicity of a drug, computer-based tools are used worldwide. The ADMET properties are strictly followed to verify the drug likeness properties for finding new drugs [36]. Compounds need to follow Lipinski's rules to be a drug. However, it is not necessary to follow Lipinski's rules. Some compounds may not follow these rules and still be effective drugs, such as natural products [37]. The tool named pkCSM (https://biosig.lab.uq.edu.au/pkcsm/prediction, accessed on October 4, 2024) was used to analyze the ADMET properties. Table 1 shows the ADMET analyses and druglike prediction of property for this significant chemical.
Table 1
Detailed ADMET screening for docking compound.
| Property | Model name | Predicted value | Unit |
|---|---|---|---|
| Absorption | Water solubility | -3.255 | Numeric (log mol/L) |
| Absorption | Caco-2 permeability | 1.296 | Numeric (log Papp in 10-6 cm/s) |
| Absorption | Intestinal absorption (Human) | 91.871 | Numeric (% Absorbed) |
| Absorption | Skin permeability | -2.735 | Numeric (log Kp) |
| Absorption | P-glycoprotein substrate | No | Categorical (Yes/No) |
| Absorption | P-glycoprotein I inhibitor | No | Categorical (Yes/No) |
| Absorption | P-glycoprotein II inhibitor | Yes | Categorical (Yes/No) |
| Distribution | VDss (Human) | -0.974 | Numeric (log L/kg) |
| Distribution | Fraction unbound (Human) | 0.146 | Numeric (Fu) |
| Distribution | BBB permeability | -0.961 | Numeric (log BB) |
| Distribution | CNS permeability | -1.176 | Numeric (log PS) |
| Metabolism | CYP2D6 substrate | No | Categorical (Yes/No) |
| Metabolism | CYP3A4 substrate | Yes | Categorical (Yes/No) |
| Metabolism | CYP1A2 inhibitor | No | Categorical (Yes/No) |
| Metabolism | CYP2C19 inhibitor | No | Categorical (Yes/No) |
| Metabolism | CYP2C9 inhibitor | No | Categorical (Yes/No) |
| Metabolism | CYP2D6 inhibito | No | Categorical (Yes/No) |
| Metabolism | CYP3A4 inhibitor | No | Categorical (Yes/No) |
| Excretion | Total clearance | 1.345 | Numeric (log mL/min/kg) |
| Excretion | Renal OCT2 substrate | No | Categorical (Yes/No) |
| Toxicity | AMES toxicity | No | Categorical (Yes/No) |
| Toxicity | Max. tolerated dose (Human) | 0.225 | Numeric (log mg/kg/day) |
| Toxicity | hERG I inhibitor | No | Categorical (Yes/No) |
| Toxicity | hERG II inhibitor | No | Categorical (Yes/No) |
| Toxicity | Oral rat acute toxicity (LD50) | 2.344 | Numeric (mol/kg) |
| Toxicity | Oral rat chronic toxicity (LOAEL) | 3.667 | Numeric (log mg/kg_ bw/day) |
| Toxicity | Hepatotoxicity | No | Categorical (Yes/No) |
| Toxicity | Skin sensitization | No | Categorical (Yes/No) |
| Toxicity | T.Pyriformis toxicity | 0.285 | Numeric (log μg/L) |
| Toxicity | Minnow toxicity | -9.793 | Numeric (log mM) |
Results
Protein-to-ligand binding
First, Table 2 shows the binding energy of CoQ10 with GSK-3β, which is -7.2 kcal/mol. The binding structure is shown more clearly in Figure 5. Figure 5A shows the 3D binding structure and Figure 5B presents the 2D diagram. In Figure 5B, we can observe that CoQ10 developed connections with seven amino acids of 1GNG, including ARG220, TYR221, LYS85, GLN185, LEU132, VAL70, and ALA83. There are four pi-alkyl bonds, two alkyl bonds, and one carbon-hydrogen bond present between the interactions of CoQ10 with GSK-3β (Figure 5B).
Table 2
Baseline characteristics of the data.
| SN | Protein name | PDB ID | Chain | Mutation | Structure weight | Binding energy Coenzyme Q10 |
|---|---|---|---|---|---|---|
| 1 | GSK-3β | 1GNG | B | No | 95.85 k DA | -7.2 kcal/mo |
| 2 | PKB | 1UNQ | A | No | 15.3 k DA | -4.6 kcal/mol |
| 3 | PI3K | 3APD | A | No | 111.02 k DA | -6.4 kcal/mo |
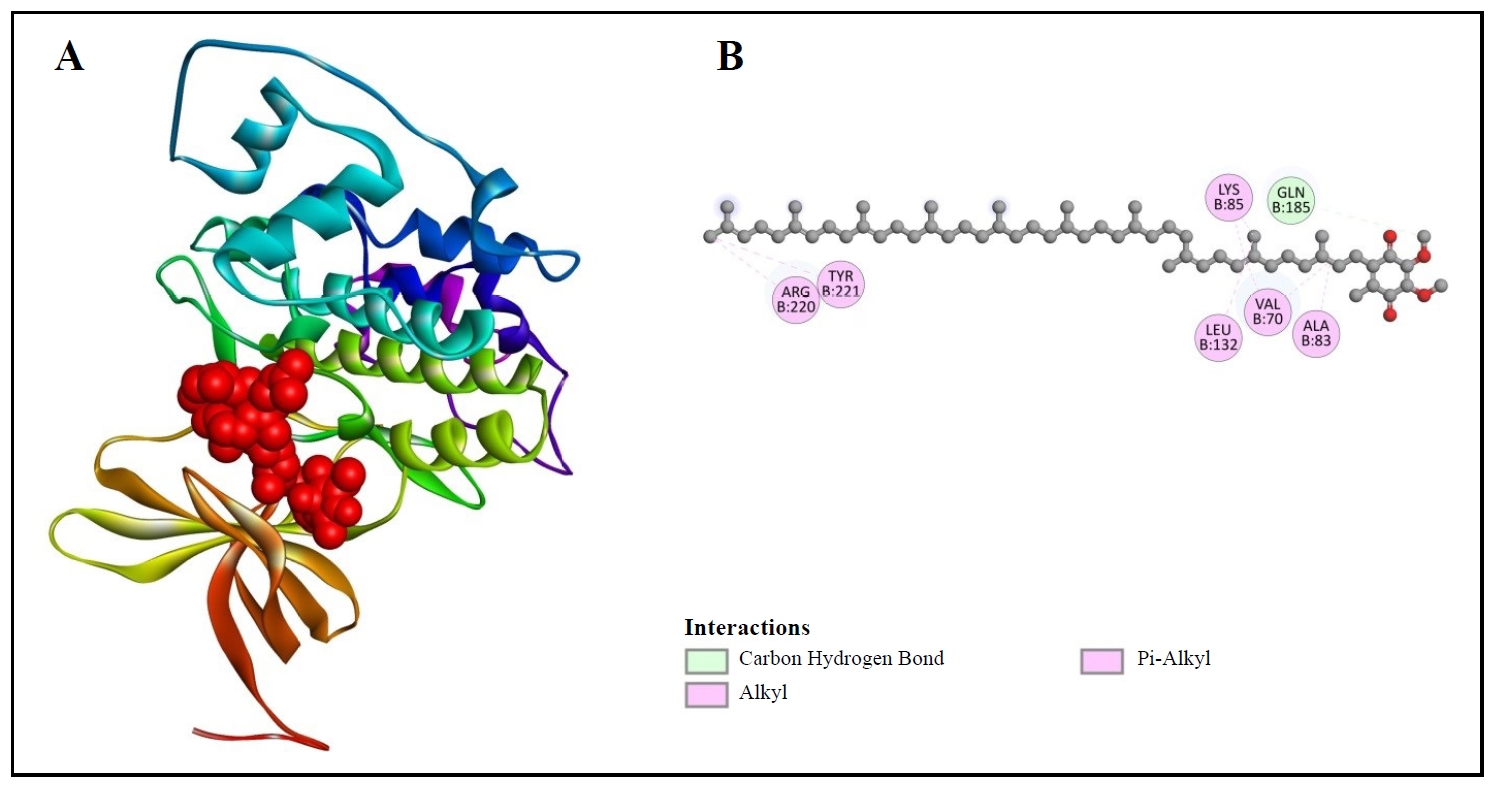
Figure 5. 3D (A) and 2D (B) structures of coenzyme Q10 binding with GSK-3β.
Furthermore, Table 2 represents the binding affinity of CoQ10 with PKB, which is -4.6 kcal/mol. We can visualize their binding site in Figure 6. Figure 6A reveals the 3D binding structure between PKB and CoQ10. Figure 6B indicates the 2D structure. GLN79, TRP80, ASN54, LEU78, THR34, SER56, ASP32, GLY33, LEU110, ALA58, GLU114, and GLN113 are the amino acids with which the compound in Figure 6B interacts. Van der Waals bonds, carbon-hydrogen bonds, alkyl, and pi-alkyl bonds are also visible in this figure.
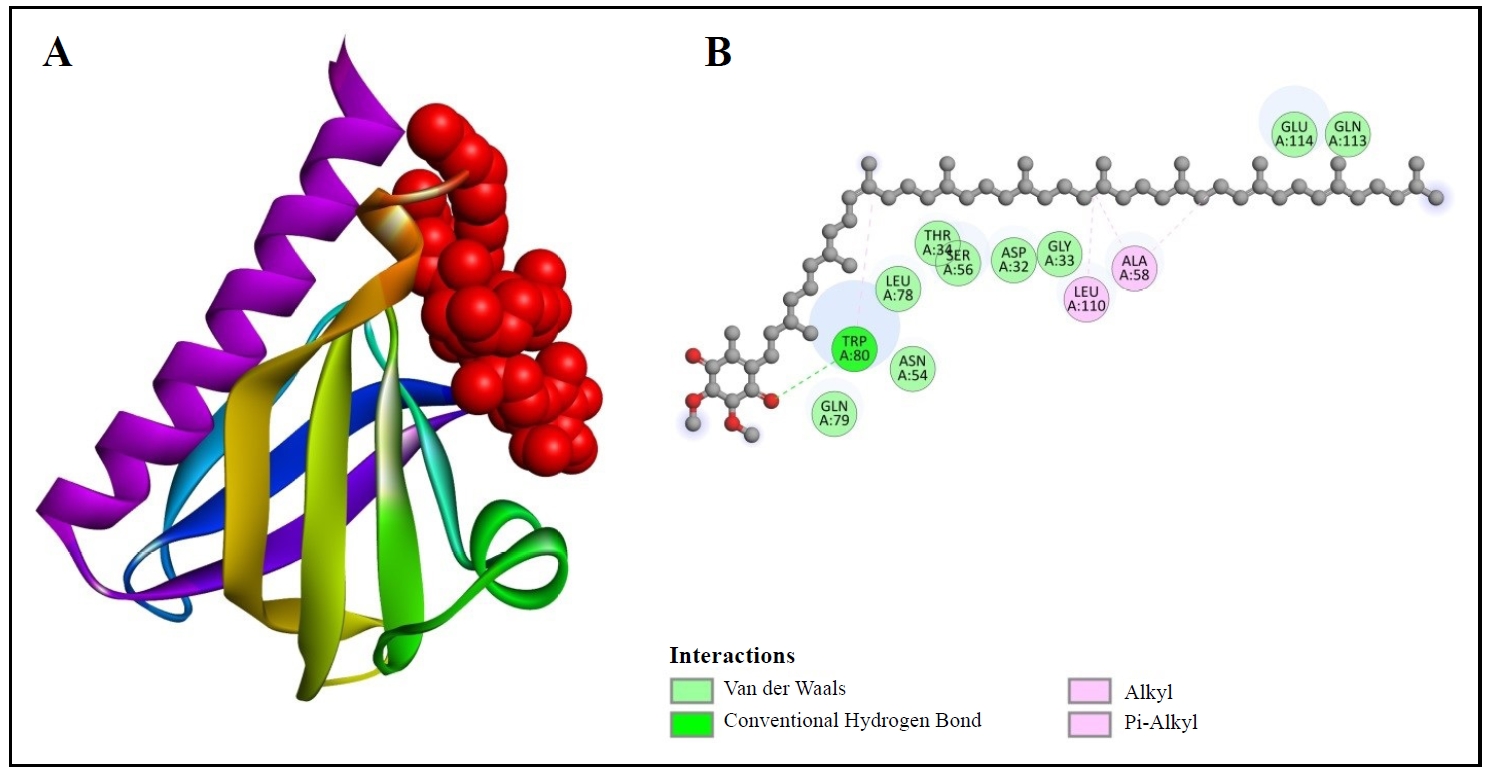
Figure 6. 3D (A) and 2D (B) structures of coenzyme Q10 binding with PKB.
Moreover, Table 2 exhibits the binding validity of CoQ10 with PI3K, which is -6.4 kcal/mol. Figure 7 provides the 3D configuration (Figure 7A) and 2D arrangement (Figure 7B) of PI3K with the chosen compound. In Figure 7B, it is noticeable that CoQ10 (compound) has developed the connections between the amino acids including, LEU823, TRP292, ARG277, HIS 295, ASP788, PRO866, and LEU865 of 3APD (protein). One conventional hydrogen bond, carbon-hydrogen bonds, alkyl, and pi-alkyl bonds are also observable in this figure.
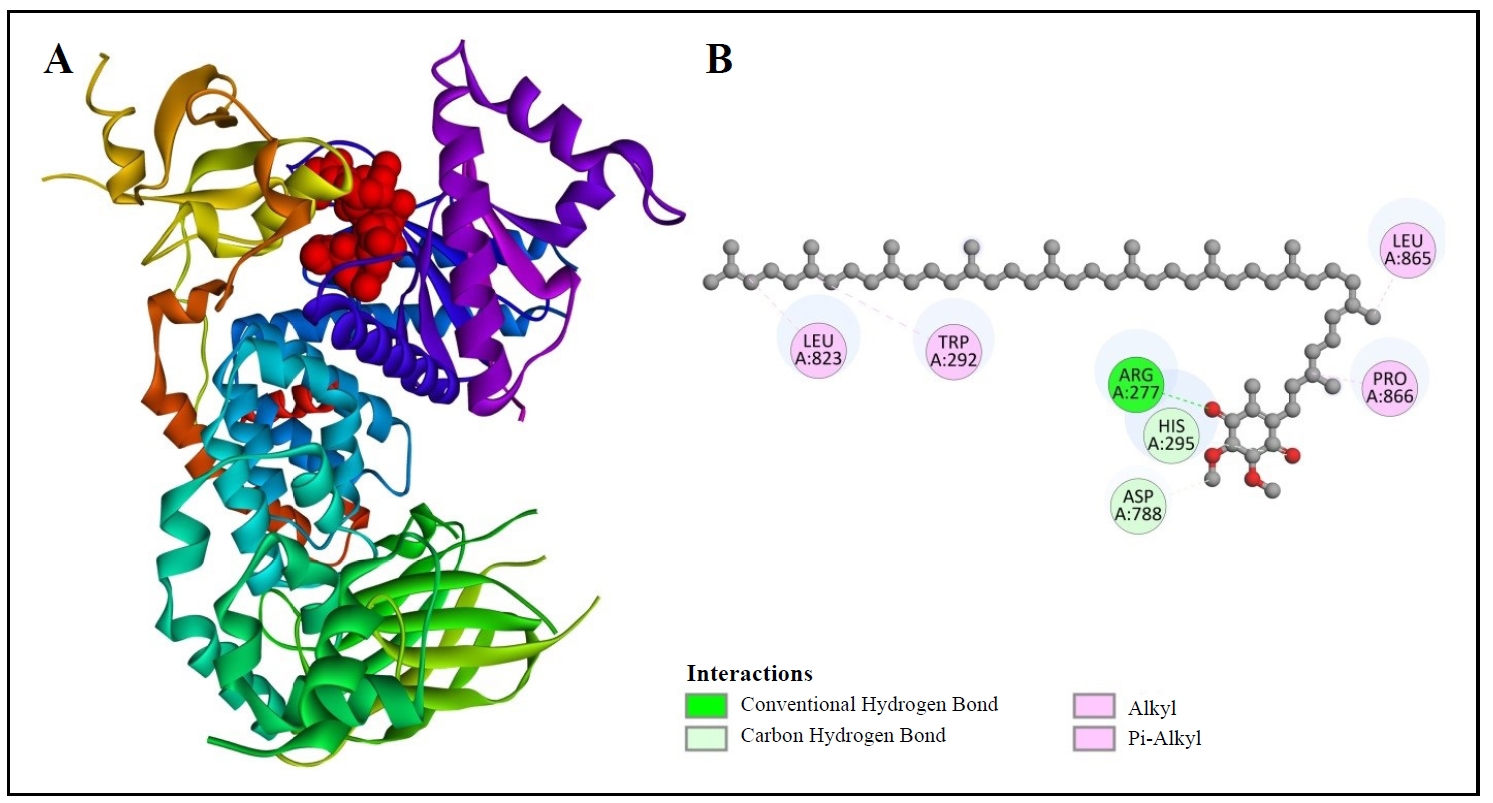
Figure 7. 3D (A) and 2D (B) structure of coenzyme Q10 binding with PI3K.
Ramachandran plot analysis
The Ramachandran plot is the graphical representation that indicates the
phi (φ) and psi (ψ) angles of the amino acids in a protein [38]. The x-axis exhibits the phi angle and the y-axis shows the psi angle
[39]. The graph indicates
distinctive regions where various phi and psi combinations are available.
The important regions are the lefthanded alpha helix, the right-handed
alpha helix, and the beta sheet.
In Figure 8, it is noticeable
that amino acid residues of all proteins are heavily represented in the
favorable regions. Besides, some residues exist outside of this region.
According to Figure 8B, amnio
acids (green colored) are clustered in the favorable regions (red
colored), which means that most of the residues are in stable
conformations, and glycine residues (purple colored) show more
flexibility. Again, Figure 8A and
8C exhibit that most of the
residues are energetically present in the favorable regions, which
indicate the stable conformation of these proteins, and glycine residues
represent flexibility.
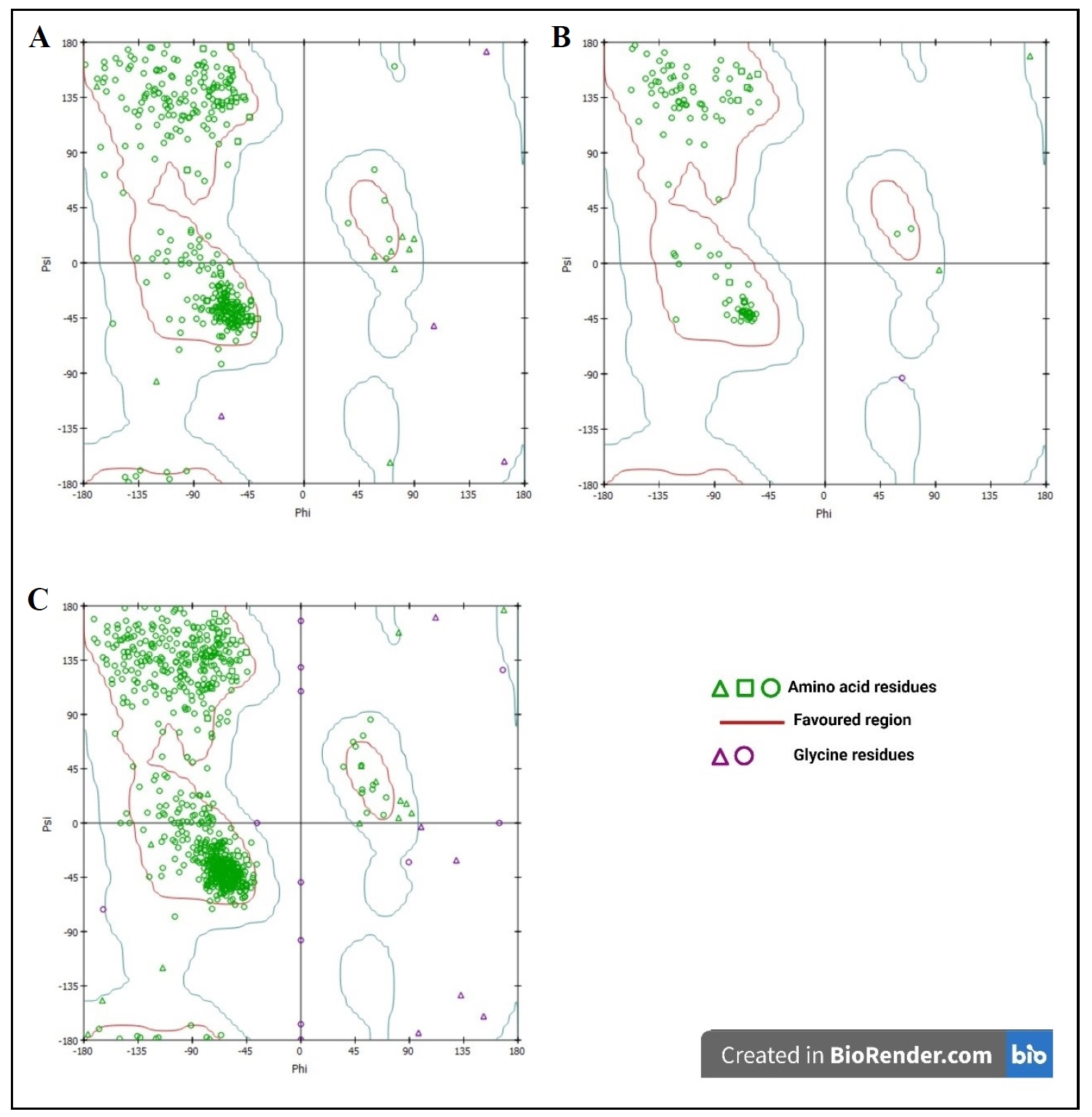
Figure 8. 3D (A) and 2D (B) structure of coenzyme Q10 binding with PI3K.
ADMET analysis for CoQ10
Absorption
According to Table 1:
1. Water solubility: This indicates the compound's solubility in water.
The negative number indicates low solubility, which may affect its
bioavailability when taken orally [40].
2. Caco-2 permeability: Finds out the rate at which a substance can enter
the Caco-2 cell monolayer. The value greater than 1.0 is considered to
have good permeability, which means the substance can be properly absorbed
through the gut [41].
3. Intestinal absorption (Human): This value indicates the percentage of a
compound that is absorbed by the intestine. The high value means excellent
absorption and is perfect for oral drugs [42].
4. Skin permeability: Displays the ability of that compound to enter into
skin. More negative value means low skin permeability, but might be ideal
for other routes [43].
5. P-glycoprotein substrate: Indicates that the compound is not an
activator for P-glycoprotein. As a result, this protein cannot pump drugs
to cells [44].
6. P-glycoprotein I inhibitor: The compound does not inhibit
P-glycoprotein I, which shows that it will reduce drug-drug interaction
[45].
7. P-glycoprotein II inhibitor: But inhibits P-glycoprotein II.
Distribution
Based on Table 1:
1. VDss (Human): The Volume of distribution at steady state (VDss)
exhibits how quickly a compound distributes throughout the body. The
negative result states low distribution, suggesting that the drug stays in
the plasma of blood [46].
2. Fraction unbound (Human): Displays the fraction of the drug that is
unbound and free for interaction with the target site. The lower value
represents the higher binding with plasma protein [47].
3. BBB permeability: Shows the ability of a compound to cross the
Blood-Brain-Barrier. The negative value represents low BBB permeability
[48].
4. CNS permeability: This value helps to identify a compound's capability
to enter into CNS tissues. Negative result is showing poor CNS activation
[49].
Metabolism
As mentioned by
Table 1:
1. CYP2D6 substrate: This result indicates that the compound cannot be
broken down by the cytochrome P450 2D6 enzyme, which limits the
possibility of certain metabolic interactions [50].
2. CYP3A4 substrate: The compound presents that it can be metabolized by
CYP3A4 enzyme. It is a very important enzyme for drug metabolism [51].
3. CYP1A2 inhibitor: The compound shows that it does not create
disturbance with the CYP1A2 enzyme. 4. CYP2C19 inhibitor: The identified
substance is not an inhibitor for CYP2C19, which means it minimizes the
drug-drug interactions [52].
5. CYP2C9 inhibitor: The compound indicates that it does not interfere in
the metabolism of drugs that are processed by the CYP2C9 enzyme.
6. CYP2D6 inhibitor: Shows no inhabitation, which means decreases the risk
of drug-drug connections.
7. CYP3A4 inhibitor: The compound does not inhibit CYP3A4.
Excretion
From the point of view of
Table 1:
1. Total clearance: The total clearance value exhibits that the compound
has a moderate clearance rate.
2. Renal OCT2 substrate: The result represents that the compound is not a
substrate for renal organic cation transporter 2. As a result, the
compound won't be affected while transporting through that pathway [53].
Toxicity
According to Table 1:
1. AMES toxicity: The compound has no AMES toxicity, which means it is not
mutagenic and will not cause any genetic mutations [54].
2. Max. tolerated dose (Human): 0.225 log mg/kg/day is the max
dose of that substance for humans. This is the safe and helpful dose level
according to the substance [55].
3. hERG I and II inhibitor: It does not inhibit hERG channels and decrease
cardiac toxicity risk [56].
4. Oral rat acute toxicity (LD50): 2.344 mol/kg is the compound's acute
toxicity level [57].
5. Oral rat chronic toxicity (LOAEL): 3.667 log mg/kg_ bw/day is the level
when the compound can create chronic toxicity [58].
6. Hepatotoxicity: It has no hepatotoxicity which results in no toxicity
in the liver and ensures the overall safety [59].
7. Skin sensitization: The compound has no skin sensitization, which
indicates it will not create an allergic reaction in the skin [60].
8. T.pyriformis toxicity: 0.285 log μg/L shows that the compound can cause
a low toxicity to T.pyriformis [61].
9. Minnow toxicity: The value is negative, which means low toxicity to
fish [62].
Discussion
The present investigation focuses on the therapeutic properties of CoQ10
to find the possibilities to eradicate AD, based on molecular docking and
ADMET analysis. The results provide significant information about the
connection between CoQ10 and the proteins that are involved in oxidative
stress and brain inflammasome [63]. These are the pathways that enhance AD. The molecular docking results
exposed links between CoQ10 and the target proteins: GSK-3β, PKB, and
PI3K.
CoQ10 shows the highest binding affinity with GSK-3β, which is -7.2
kcal/mol, and has developed connections with seven amino acids, including
ARG220, TYR221, and LYS85. This strong interaction presents that CoQ10 can
affect GSK-3β activity, which is crucial for tau protein phosphorylation.
Furthermore, the binding affinity of CoQ10 with PI3K is -6.4 kcal/mol with
the interactions between the compound and amino acids such as LEU823,
TRP292, and ARG277. These interactions with PI3K are involved in the
PI3K/Akt signaling pathway, which plays a significant role in
neuroprotection and cell survival. Additionally, CoQ10 exhibits the lower
binding affinity with PKB, which is -4.6 kcal/mol, and the compound forms
connections with various amino acids, including GLN79, TRP80, and ASN54.
These interactions may contribute to the PI3K/Akt pathway, potentially
increasing the neuroprotective effects. Based on previous studies, these
molecular interactions represent the mechanistic basis for the CoQ10
effects on antioxidant and neuroprotection [13-15]. The capability of CoQ10
to interact with these proteins highlights the compound's possibility in
reducing oxidative stress and brain inflammation [64].
The ADMET analysis of CoQ10 revealed several favorable pharmacokinetic and
toxicological properties [65]:
1. Absorption: CoQ10 expresses high intestinal absorption (91.871%) and
good Caco-2 permeability (1.296), indicating excellent oral
bioavailability. But it has low water solubility (-3.255 log mol/L), which
may create challenges for formulation and delivery [40-42].
2. Distribution: The negative BBB permeability (-0.961) and CNS
permeability (1.176) results present limited entrance into the central
nervous system. This is a limitation for treating neurological disorders
like AD [46-48].
3. Metabolism: CoQ10 is a substrate for CYP3A4 but not for CYP2D6.
Further, it does not inhibit major CYP enzymes. These properties show the
lower risk of drug-drug interactions. It is valuable for a patient who
takes multiple drugs at a time [66].
4. Excretion: The compound showed a medium clearance rate, which is 1.345
log mL/min/kg, which indicates a balanced elimination profile.
5. Toxicity: CoQ10 published a favorable toxicity profile with no AMES
toxicity, hepatotoxicity, or skin sensitization. It also showed no
inhibition of hERG channels, which reduces the risk of cardiotoxicity. The
maximum tolerated dose and chronic toxicity levels mean that it is safe
for human use [54-56].
These ADMET properties establish the safety profile for CoQ10 in clinical
use and support its potential for longterm use in chronic conditions like
AD.
The proper binding affinities between CoQ10 and the key proteins involved
in AD pathology and its positive ADMET result encourage further
investigation on CoQ10 for its potential therapeutic properties for
treating AD. The interactions with GSK-3β show a better result that
indicates CoQ10 may help in reducing tau protein phosphorylation, a
hallmark of AD [19].
However, the poor BBB permeability of CoQ10 presents a significant
challenge that needs to be focused on. Future research should place
importance on developing the delivery systems and chemical modifications
to enhance the CNS penetration of CoQ10 to increase its beneficial
properties.
Additionally, this in silico study provides valuable data [67]. This information needs to be identified through in vitro and
in vivo experiments. More studies are needed to find out the
impacts of CoQ10 on tau phosphorylation, amyloid-β plaque formation, and
neuroinflammation.
Conclusions
This study demonstrates the therapeutic potential of CoQ10 in mitigating
key pathological mechanisms associated with AD, such as oxidative stress
and neuroinflammation. Molecular docking results revealed strong
interactions between CoQ10 and GSK-3β, PKB, and PI3K, suggesting that
CoQ10 may reduce tau phosphorylation and enhance neuroprotection via the
PI3K/Akt signaling pathway. ADMET analysis confirmed CoQ10's favorable
pharmacokinetic profile, including high intestinal absorption and low
toxicity, though its limited blood-brain barrier permeability remains a
challenge for treating central nervous system disorders like AD.
Overall, this in silico investigation supports CoQ10 as a
promising candidate for therapeutic development in AD. However, further
experimental studies, including in vitro and
in vivo models, are required to validate its efficacy in reducing
amyloid-β plaques, tau tangles, and overall disease progression. Future
research should also explore strategies to enhance CoQ10 delivery to the
brain for more effective treatment outcomes.
Declarations
Availability of data and materials
Not applicable.
Financial support and sponsorship
None.
Conflicts of interest
Not applicable.
References
1. Kumar A, Sidhu J, Lui F, & Tsao JW. Alzheimer disease. In: StatPearls. 2024, edn.
2. Holtzman DM, Morris JC, & Goate AM. Alzheimer's disease: the challenge of the second century. Sci Transl Med, 2011, 3(77): 77sr71. [Crossref]
3. Vaiserman A, Koliada A, & Lushchak O. Neuroinflammation in pathogenesis of Alzheimer's disease: phytochemicals as potential therapeutics. Mech Ageing Dev, 2020, 189: 111259. [Crossref]
4. Adamu A, Li S, Gao F, & Xue G. The role of neuroinflammation in neurodegenerative diseases: current understanding and future therapeutic targets. Front Aging Neurosci, 2024, 16: 1347987. [Crossref]
5. Selkoe DJ, & Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol Med, 2016, 8(6): 595- 608. [Crossref]
6. Wang X, Wang W, Li L, Perry G, Lee HG, & Zhu X. Oxidative stress and mitochondrial dysfunction in Alzheimer's disease. Biochim Biophys Acta, 2014, 1842(8): 1240- 1247. [Crossref]
7. Wyss-Coray T, & Rogers J. Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring Harb Perspect Med, 2012, 2(1): a006346. [Crossref]
8. Beurel E, Grieco SF, & Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther, 2015, 148: 114-131. [Crossref]
9. Beal MF. Coenzyme Q10 administration and its potential for treatment of neurodegenerative diseases. Biofactors, 1999, 9(2-4): 261-266. [Crossref]
10. Butterfield DA, & Halliwell B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease.Nat Rev Neurosci, 2019, 20(3): 148-160. [Crossref]
11. Zhang B, Li H, Yu K, & Jin Z. Molecular docking-based computational platform for high-throughput virtual screening. CCF Trans High Perform Comput, 2022, 4(1): 63-74. [Crossref]
12. Scarpelli M, Todeschini A, Rinaldi F, Rota S, Padovani A, & Filosto M. Strategies for treating mitochondrial disorders: an update. Mol Genet Metab, 2014, 113(4): 253- 260. [Crossref]
13. Mantle D, & Dybring A. Bioavailability of coenzyme Q(10): an overview of the absorption process and subsequent metabolism. Antioxidants (Basel), 2020, 9(5): 386- 396. [Crossref]
14. Calabrese V, Scapagnini G, Colombrita C, Ravagna A, Pennisi G, Giuffrida Stella AM, et al. Redox regulation of heat shock protein expression in aging and neurodegenerative disorders associated with oxidative stress: a nutritional approach. Amino Acids, 2003, 25(3-4): 437-444. [Crossref]
15. Jiménez-Jiménez FJ, Alonso-Navarro H, García-Martín E, & Agúndez JAG. Coenzyme Q10 and dementia: a systematic review. Antioxidants (Basel), 2023, 12(2): 533-543. [Crossref]
16. Chang PS, Chou HH, Lai TJ, Yen CH, Pan JC, & Lin PT. Investigation of coenzyme Q10 status, serum amyloid-β, and tau protein in patients with dementia. Front Aging Neurosci, 2022, 14: 910289. [Crossref]
17. Bagheri S, Haddadi R, Saki S, Kourosh-Arami M, Rashno M, Mojaver A, et al. Neuroprotective effects of coenzyme Q10 on neurological diseases: a review article. Front Neurosci, 2023, 17: 1188839. [Crossref]
18. Rauchová H. Coenzyme Q10 effects in neurological diseases. Physiol Res, 2021, 70(Suppl4): S683-s714. [Crossref]
19. Abuelezz SA, & Hendawy N. Spotlight on coenzyme Q10 in scopolamine-induced Alzheimer's disease: oxidative stress/PI3K/AKT/GSK 3ß/CREB/BDNF/TrKB. J Pharm Pharmacol, 2023, 75(8): 1119-1129. [Crossref]
20. Fišar Z, & Hroudová J. CoQ(10) and mitochondrial dysfunction in Alzheimer's disease. Antioxidants (Basel), 2024, 13(2): 191-203. [Crossref]
21. Vázquez-Jiménez LK, Juárez-Saldivar A, Gómez-Escobedo R, Delgado-Maldonado T, Méndez-Álvarez D, Palos I, et al. Ligand-based virtual screening and molecular docking of benzimidazoles as potential inhibitors of triosephosphate isomerase identified new trypanocidal agents. Int J Mol Sci, 2022, 23(17):10047. [Crossref]
22. O'Boyle NM, Morley C, & Hutchison GR. Pybel: a Python wrapper for the openbabel cheminformatics toolkit. Chem Cent J, 2008, 2: 5-14. [Crossref]
23. Laskowski RA, MacArthur MW, & Thornton JM. Validation of protein models derived from experiment. Curr Opin Struct Biol, 1998, 8(5): 631-639. [Crossref]
24. Colovos C, & Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci, 1993, 2(9): 1511-1519. [Crossref]
25. Wiederstein M, & Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res, 2007, 35(Web Server issue): W407-410. [Crossref]
26. A. Yakubu, Donato MD, & Imumorin IG. Modelling functional and structural impact of non-synonymous single nucleotide polymorphisms of the DQA1 gene of three Nigerian goat breeds. S Afr J Anim Sci, 2017, 47(2): 146– 156. [Crossref]
27. Kwofie SK, Dankwa B, Odame EA, Agamah FE, Doe LPA, Teye J, et al. In silico screening of isocitrate lyase for novel anti-buruli ulcer natural products originating from Africa. Molecules, 2018, 23(7): 1550-1560. [Crossref]
28. Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci, 2021, 30(1): 70-82. [Crossref]
29. Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci, 2018, 27(1): 14-25. [Crossref]
30. Wang S, Jiang JH, Li RY, & Deng P. Docking-based virtual screening of TβR1 inhibitors: evaluation of pose prediction and scoring functions. BMC Chem, 2020, 14(1): 52- 62. [Crossref]
31. Alhawarri MB, Dianita R, Rawa MSA, Nogawa T, & Wahab HA. Potential anti-cholinesterase activity of bioactive compounds extracted from Cassia grandis L.f. and Cassia timoriensis DC. Plants (Basel), 2023, 12(2): 344-354. [Crossref]
32. McCullagh M, Zeczycki TN, Kariyawasam CS, Durie CL, Halkidis K, Fitzkee NC, et al. What is allosteric regulation? Exploring the exceptions that prove the rule! J Biol Chem, 2024, 300(3): 105672. [Crossref]
33. Pawar RP, & Rohane SH. Role of autodock vina in PyRx molecular docking. J Res Chem, 2021, 14(2): 132–134. [Crossref]
34. E. López-Camacho, M. J. García-Godoy, J. García-Nieto, A. J. Nebro, & J. F. Aldana-Montes. Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics). 2016, 97(02): 65-77. [Crossref]
35. Mir WR, Bhat BA, Rather MA, Muzamil S, Almilaibary A, Alkhanani M, et al. Molecular docking analysis and evaluation of the antimicrobial properties of the constituents of Geranium wallichianum D. Don ex Sweet from Kashmir Himalaya. Sci Rep, 2022, 12(1): 12547. [Crossref]
36. Daoud NE, Borah P, Deb PK, Venugopala KN, Hourani W, Alzweiri M, et al. ADMET profiling in drug discovery and development: perspectives of In silico, in vitro and integrated approaches. Curr Drug Metab, 2021, 22(7): 503- 522. [Crossref]
37. V. Ivanović, M. Rančić, B. Arsić, & A. Pavlović. Lipinski's rule of five, famous extensions and famous exceptions. Chemia Naissensis, 2020, 3(1): 171–181. [Crossref]
38. Aarthy M, & Singh SK. Envisaging the conformational space of proteins by coupling machine learning and molecular dynamics. Adv Protein Mol Struct Biol Metho, 2022: 467–475. [Crossref]
39. Priestle JP, & Paris CG. Experimental techniques and data banks. Guidebook on Molecular Modeling in Drug Design, 1996: 139–217. [Crossref]
40. Stielow M, Witczyńska A, Kubryń N, Fijałkowski Ł, Nowaczyk J, & Nowaczyk A. The bioavailability of drugsthe current state of knowledge. Molecules, 2023, 28(24): 8038-8048. [Crossref]
41. Kus M, Ibragimow I, & Piotrowska-Kempisty H. Caco-2 cell line standardization with pharmaceutical requirements and in vitro model suitability for permeability assays. Pharmaceutics, 2023, 15(11): 2523-2533. [Crossref]
42. Van Den Abeele J, Rubbens J, Brouwers J, & Augustijns P. The dynamic gastric environment and its impact on drug and formulation behaviour.Eur J Pharm Sci, 2017, 96: 207-231. [Crossref]
43. Neupane R, Boddu SHS, Renukuntla J, Babu RJ, & Tiwari AK. Alternatives to biological skin in permeation studies: current trends and possibilities. Pharmaceutics, 2020, 12(2): 152-162. [Crossref]
44. Nguyen TT, Duong VA, & Maeng HJ. Pharmaceutical formulations with p-glycoprotein inhibitory effect as promising approaches for enhancing oral drug absorption and bioavailability. Pharmaceutics, 2021, 13(7): 1103-1113. [Crossref]
45. Callaghan R, Luk F, & Bebawy M. Inhibition of the multidrug resistance P-glycoprotein: time for a change of strategy? Drug Metab Dispos, 2014, 42(4): 623-631. [Crossref]
46. Holford N, & Yim DS. Volume of distribution. Transl Clin Pharmacol, 2023, 24: 74-77. [Crossref]
47. Dasgupta A. Monitoring free mycophenolic acid concentration: is there any clinical advantage? 2016: 83-107.
48. Nau R, Sörgel F, & Eiffert H. Penetration of drugs through the blood-cerebrospinal fluid/blood-brain barrier for treatment of central nervous system infections. Clin Microbiol Rev, 2010, 23(4): 858-883. [Crossref]
49. Carpenter TS, Kirshner DA, Lau EY, Wong SE, Nilmeier JP, & Lightstone FC. A method to predict blood-brain barrier permeability of drug-like compounds using molecular dynamics simulations. Biophys J, 2014, 107(3): 630-641. [Crossref]
50. Taylor C, Crosby I, Yip V, Maguire P, Pirmohamed M, & Turner RM. A Review of the important role of CYP2D6 in pharmacogenomics. Genes (Basel), 2020, 11(11): 1295- 1302. [Crossref]
51. Lee J, Beers JL, Geffert RM, & Jackson KD. A review of CYP-mediated drug interactions: mechanisms and in vitro drug-drug interaction assessment. Biomolecules, 2024, 14(1): 99-101. [Crossref]
52. Shubbar Q, Alchakee A, Issa KW, Adi AJ, Shorbagi AI, & Saber-Ayad M. From genes to drugs: CYP2C19 and pharmacogenetics in clinical practice. Front Pharmacol, 2024, 15: 1326776. [Crossref]
53. Hacker K, Maas R, Kornhuber J, Fromm MF, & Zolk O. Substrate-dependent inhibition of the human organic cation transporter OCT2: a comparison of metformin with experimental substrates. PLoS One, 2015, 10(9): e0136451. [Crossref]
54. Thomas DN, Wills JW, Tracey H, Baldwin SJ, Burman M, Williams AN, et al. Ames test study designs for nitrosamine mutagenicity testing: qualitative and quantitative analysis of key assay parameters. Mutagenesis, 2024, 39(2): 78-95. [Crossref]
55. Gad SC. Maximum tolerated dose. In: Encyclopedia of Toxicology (Fourth Edition). 2024, edn. Academic Press.
56. Stergiopoulos C, Tsopelas F, & Valko K. Prediction of hERG inhibition of drug discovery compounds using biomimetic HPLC measurements. Admet dmpk, 2021, 9(3): 191-207. [Crossref]
57. Niyomchan A, Chatgat W, Chatawatee B, Keereekoch T, Issuriya A, Jaisamut P, et al. Safety evaluation of the polyherbal formulation nawatab: acute and subacute oral toxicity studies in rats. Evid Based Complement Alternat Med, 2023, 2023: 9413458. [Crossref]
58. Selvestrel G, Lavado GJ, Toropova AP, Toropov AA, Gadaleta D, Marzo M, et al. Monte carlo models for subchronic repeated-dose toxicity: systemic and organspecific toxicity. Int J Mol Sci, 2022, 23(12): 6615-6625. [Crossref]
59. Björnsson ES. Hepatotoxicity by drugs: the most common implicated agents. Int J Mol Sci, 2016, 17(2): 224- 234. [Crossref]
60. Bialas I, Zelent-Kraciuk S, & Jurowski K. The skin sensitisation of cosmetic ingredients: review of actual regulatory status. Toxics, 2023, 11(4): 392-404. [Crossref]
61. Luan F, Wang T, Tang L, Zhang S, & Cordeiro M. Estimation of the toxicity of different substituted aromatic compounds to the aquatic ciliate Tetrahymena pyriformis by QSAR approach. Molecules, 2018, 23(5): 1002-1013. [Crossref]
62. Wu X, Zhang Q, & Hu J. QSAR study of the acute toxicity to fathead minnow based on a large dataset. SAR QSAR Environ Res, 2016, 27(2): 147-164. [Crossref]
63. Dumont M, Kipiani K, Yu F, Wille E, Katz M, Calingasan NY, et al. Coenzyme Q10 decreases amyloid pathology and improves behavior in a transgenic mouse model of Alzheimer's disease. J Alzheimers Dis, 2011, 27(1): 211- 223. [Crossref]
64. Hernández-Camacho JD, Bernier M, López-Lluch G, & Navas P. Coenzyme Q(10) supplementation in aging and disease. Front Physiol, 2018, 9: 44-54. [Crossref]
65. Zhu ZG, Sun MX, Zhang WL, Wang WW, Jin YM, & Xie CL. The efficacy and safety of coenzyme Q10 in Parkinson's disease: a meta-analysis of randomized controlled trials. Neurol Sci, 2017, 38(2): 215-224. [Crossref]
66. Turunen M, Olsson J, & Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta, 2004, 1660(1- 2): 171-199. [Crossref]
67. Kitchen DB, Decornez H, Furr JR, & Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov, 2004, 3(11): 935-949. [Crossref]