Open Access | Mini Review
This work is licensed under a Creative
Commons Attribution-ShareAlike 4.0 International License.
Human microbiota alterations — emerging predictors of renal diseases and kidney-specific aging
# These authors contributed equally to this work.
* Corresponding author: Chao Zhao
Mailing address: MOE/NHC/CAMS Key Lab of Medical Molecular Virology, School of Basic Medical Sciences & National
Clinical Research Center for Aging and Medicine, Huashan Hospital, Shanghai Medical College, Fudan University, Shanghai,
200032, China.
Email: czhao@fudan.edu.cn
Received: 24 November 2022 / Revised: 12 January 2023 / Accepted: 14 February 2023 / Published: 29 March 2023
DOI: 10.31491/APT.2023.03.105
Abstract
Rapid advances in sequencing and analytical technologies have increased our understanding of the interactions between the microbiome and the host. The microbiome plays a variety of physiological roles in the health of the host and pathological roles in disease; for example, the microbiome changes significantly when kidney health is compromised and during kidney-specific aging. At present, good diagnostic markers are absent for early renal injury. However, the composition of the microbiome could provide useful indications for disease diagnosis and treatment beyond more conventional diagnostic markers. Such indications are well represented in chronic kidney disease, immunoglobulin A nephropathy, idiopathic nephrotic syndrome, and diabetic nephropathy. With sensitivity, specificity, and stability, the microbiota can provide more possibilities for the diagnosis of the early recognition of asymptomatic renal injury and aging. Moreover, probiotics and microbial metabolites have shown beneficial effects in the treatment of kidney diseases. Therapeutic measures targeting the microbiota can effectively improve the immune response and inflammatory state of the body. This paper reviews the current evidence on how the microbiome is not only a potentially effective tool for clinical diagnosis but also an important focus for the study of kidney disease and aging.
Keywords
Human microbiota, kidney-specific aging, renal disease, high-throughput sequencing, predictor
Introduction
The development of sequencing technology has shown that—despite their small size—microbes play a variety of crucial roles in the health of the host, as well as the pathogenesis of multiple diseases. In addition to their cellular functions, microbes are now known to constitute an important part of human organs, tissues, and systems [1]. There are at least ten times as many bacteria as human cells in the body [2]. The symbiotic microbiome of the human body is intimately connected to host physiology. Consequently, microbes also play important roles in human health and disease. The symbiotic microbiome can be significantly altered by lifestyle, diet, and even exercise [3, 4], and changes in the microbiome have been shown to play a role in the pathophysiological processes of many diseases [5]. Therefore, the interaction between human symbiotic microorganisms and the body may be even far more complex than imagined.
The symbiotic microbiome’s new identity in human disease
The human microbiome has been extensively analyzed
in different states in recent years. It is recognized that,
when the host is in an abnormal state, it will interact with
and induce changes in the symbiotic microbiome. In the
healthy state, the human microbiome fluctuates but is generally quite stable. Organ-specific community structures
exist; for example, skin microbes have distinct community
characteristics in different parts of the skin [6]. Oral microbes also differ from person to person [7]. The intestinal
tract, with the highest microbial load, is also relatively
stable [8]. Although the intestinal flora can be temporarily changed by diarrhea, antibiotic consumption, or other
influences, the original floral structure reappears after a
period of recovery [9]. Thus, even when the organism is
in a state of disease, the microbiome also possesses certain stable characteristics. The microbiome and systemic
metabolism, endocrine and immune systems have systemic effects at the host level. Microorganisms can often
respond to subtle changes in the host under abnormal conditions, thus alterations to the microbiome may potentially
have diagnostic or prognostic value.
Evaluation of the microbiota may be useful as a noninvasive method for diagnostic purposes. The sensitivity
of the microbial community structure to abnormal states
may hold potential as new markers of disease that could
complement the traditional analysis of body fluid samples,
tissue sections, and other clinical methods. The diagnostic
potential of the microbiota has been identified in many
diseases. For example, changes in the characteristics of
the intestinal microflora can potentially predict early lung
cancer [10]. Moreover, changes in the intestinal microbiome are related to the severity of coronary artery disease
[11], and other changes in microbial diversity are directly
and indirectly associated with hypertension [12]. In addition, studies have shown that gut microbiome alterations
predispose to numerous neurological diseases [13]. Therefore, charting of the microbial map of the microbiome
may significantly contribute to the diagnosis and targeted
treatment of a wide range of diseases.
The complex role of the microbiome in the aging process
of the human body is gradually being elucidated with
the advancement of understanding. The composition and
structure of human microorganisms are constantly changing with age [14]. This age-related perturbation is accompanied by the occurrence of states such as inflammation,
which largely influences the appearance of age-related
pathological states [15]. Thus, interactions between the
human microbiome and the host largely influence the rate
of aging. Microbiome-specific modulation becomes an
important part of anti-aging research [16]. The specific
gut microbial composition has also been suggested as a
predictor of aging [17]. Adequate elaboration of the role
of microbiota in the aging process would be very useful
in the regulation of the aging process and the response to
diseases of aging.
Kidney aging is one of the important aspects of systemic
aging. The physiological structure and function of the
kidney become damaged during the aging process, and
this damage can result in a series of pathological processes and diseases [18]. Renal disease is an increasingly
important global public health problem [19]. Due to the
lack of obvious clinical manifestations in the early stages
of kidney disease, most patients have developed the latestage disease by the time they are diagnosed and thus
have a poor prognosis. Therefore, the discovery of new
therapeutic markers and targets for kidney disease is very
important [20]. The development of high-throughput sequencing technology and the emergence of databases can
help to better understand the relationship between diseases
and microorganisms. The strong associations between gut
microbiota alterations and kidney disease have also been
extensively explored. A high abundance of microbiota in
patients with kidney disease can distinguish illness (Figure 1). High-throughput studies based on renal pathological
status and microbiome also confirmed this complex association (Figure 2). As a result of the increasing attention
being paid to the complex relationship between microorganisms and diseases in recent years, microorganisms
have become a new target in the etiology and clinical diagnosis of renal function injury.
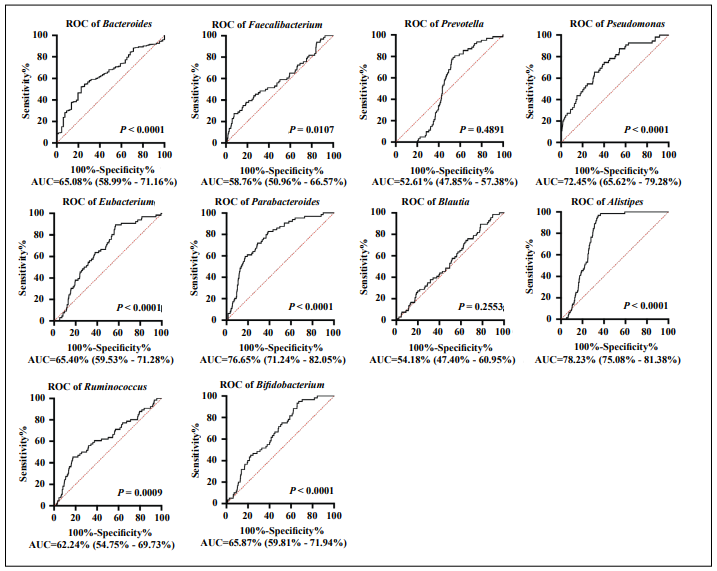
Figure 1. Receiver operating characteristic (ROC) curves of the top ten genera of gut microbiota associated with kidney disease. ROC curves of the top ten microbial genera with the highest relative abundance in kidney disease in the gut microbiome database GMrepo (https://gmrepo.humangut. info/phenotypes/); the area under the curve (AUC) is shown for each genus. Gut microbiota shows predictive ability in differentiating patients with kidney disease (P > 0.05, AUC > 0.5).
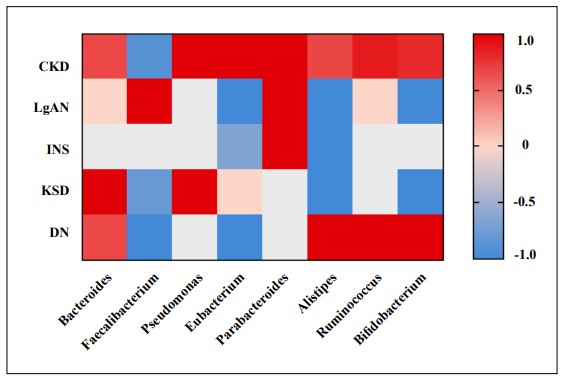
Figure 2. Heatmap of the associations between alterations to specific genera of the gut microbiota and various kidney diseases. Literature statistics on the relative abundance of specific microbiota in kidney disease and health groups compared. Red indicates a higher proportion of literature with a high relative abundance of specific microbiota in kidney disease; blue indicates a higher proportion of literature with a low relative abundance of specific microbiota in kidney disease. Zero indicates that the genus does not differ between healthy controls and patients with the disease in recent reports. Colorless patches suggest that no studies found associated microorganisms are detected in the gut microbiome of patients. Detailed data are provided in Supplementary Tables 1 and 2. (CKD: Chronic kidney disease, IgAN: Immunoglobulin A nephropathy, INS: Idiopathic nephrotic syndrome, DN: Diabetic nephropathy).
Microbiome alterations can distinguish between renal physiological and pathophysiological states
Chronic kidney disease
Chronic kidney disease (CKD) is defined as a persistent
decline in kidney function, with a glomerular filtration
rate below 60 mL/min/1.73 m2
and/or markers of kidney
damage that persist for at least three months [21]. The
clinical diagnosis of CKD is mainly determined based
on the glomerular filtration rate and serum creatinine
and albumin levels. The interpretation of these markers
is complex due to the complex etiology of nephropathy;
for example, 30% of patients with diabetic nephropathy
do not have abnormal urinary albumin levels, and serum
creatinine does not increase until at least 40% of the renal
parenchyma is damaged [22, 23]. Early diagnosis of CKD
can enable patients to receive treatment to slow down the
progression of the disease and improve their prognosis;
however, standard laboratory markers of renal deterioration are virtually unaffected in the early stages of kidney
disease. Therefore, more useful markers to evaluate the
occurrence and development of nephropathy urgently
need to be identified.
Recent advancements in multiple omics techniques have
broadened the search for such biomarkers, and the intestinal flora has been identified to play an important role in
the pathophysiology of CKD. Evidence is accumulating
that changes in the characteristics of the intestinal flora
may be clinically useful for the early identification of
CKD. Patients with CKD have lower gut microbiota diversity than healthy controls [20] and have a lower total
number of bacteria in their feces. Moreover, patients with
CKD have a lower abundance of probiotic-producing microbiota, such as Lactobacillus and Prevotella [24], and
significant enrichment of some opportunistic pathogens,
such as Actinomycetes and Proteobacteria [25]. Several
microbial markers of diagnostic significance have also
been identified. Ruminococcus and Roseburia can distinguish patients with CKD from healthy controls [26] and
patients with CKD were found to have a significantly
lower abundance of Roseburia, Faecalibacterium, and
Clostridium and significantly increased abundance of
Klebsiella and Akkermansia [20, 24]. In addition,Bacteroides eggerthii, Cetobacterium somerae, and Candidatus
Stoquefichus sp. KLE1796 can better distinguish early
CKD from traditional biochemical markers. Bacteroides eggerthii, in particular, showed good diagnostic specificity
for CKD, both alone and in conjunction with other alterations to the microbiome [27]. A core microbiome associated with the course of CKD was identified, consisting of nine genera (Escherichia_shigella, Dialister, Lachnospiraceae _ND3007_group, Pseudobutyrivibrio, Roseburia,
Paraprevotella, Ruminiclostridium, Collinsella stercoris,
and Bacteroides eggerthii). In particular, Paraprevotella,
Pseudobutyrivibrio, and Collinsella stercoris more accurately identified CKD than the classic measure of urinary
protein/creatinine. These microbial markers are highly
stable, even in the early stages of the disease [28]. Moreover, butyrate production by Roseburia inulinivorans and
Ruminococcus is significantly reduced in the early stages
of CKD [29]. These characteristic changes suggest that
changes in the microbiome in CKD patients can be an early indicator of an unhealthy state of the organism. CKD
is an important component of aging-related diseases. The
ability of microbial markers to accurately identify clinical
symptoms before they appear will further improve clinical
outcomes in aging-related diseases.
Immunoglobulin A nephropathy
Immunoglobulin A nephropathy (IgAN), the most common type of primary glomerular disease worldwide and
the leading cause of end-stage renal disease in adults has
become an important global health problem [30, 31]. Diagnosis of this disease requires a renal biopsy to examine
the deposition of immune complexes in the mesangium.
This invasive procedure can lead to kidney inflammation
and failure [32]. Although IgAN has distinct clinical features, some patients do not show significant symptoms due
to rapid changes during the disease course [33]. Therefore,
less invasive, more accurate markers are urgently needed
for the diagnosis of IgAN.
IgA regulates symbiotic bacterial homeostasis in the body
and the intestinal flora play an important role in maintaining intestinal immune stability [34]. However, many studies have shown that intestinal mucosal immune responses
related to intestinal floral disorder promote the development of IgAN [35, 36]. Therefore, microbial characteristics may be potentially useful for the diagnosis of IgAN. A
comparison of patients with advanced and non-advanced
IgAN found microbial diversity was reduced in patients
with advanced IgAN. Patients with both advanced and
non-advanced IgAN had fewer types of Bifidobacteria
than healthy subjects. Moreover, Enterococcus and Lactobacillus were reduced in patients with IgAN, and Rumencoccus, Eubacter, and Streptococcus were most abundant
in patients with advanced IgAN [37]. Compared to a
healthy control group, the abundance of Fusobacteria,
Escherichia-Shigella, Hungatella, and Eggerthella was
increased in patients with IgAN; these bacteria have a certain pathogenic potential. Moreover, Escherichia-Shigella
was negatively correlated with the estimated glomerular
filtration rate [38, 39], and Legionella, Escherichia-Shigella, and Ruminococcus were also enriched in the blood
of patients with IgAN [40]. In addition, a relatively recent
study showed that an abnormal mucosal immune response
to the anaerobic flora of the tonsils (mainly Bacteroidetes)
was related to the pathophysiology of IgAN [41]. Thus,
the decrease in probiotics and the increase in pathogenic
bacteria, and the resulting disturbance of the intestinal
microbiota may be an important part of the pathological
process of IgAN. Microbial disorders and the resulting
immune activation can be a breakthrough in disease diagnosis and clinical treatment.
Idiopathic nephrotic syndrome
Idiopathic nephrotic syndrome (INS) is a common form of podocytosis and the most common glomerular disease in children [42]. The main pathological findings include minimal change disease and focal segmental glomerulosclerosis. The clinical manifestations of INS include glomerular filtration disorder and proteinuria [43]. Compared with healthy children, the proportion of butyric acidproducing bacteria is decreased significantly in the intestines of children with INS [44], and metagenomic analysis confirmed this result [45]. Characteristic changes in the intestinal flora were also observed in adult patients with INS. The bacterial diversity of patients was significantly altered compared to healthy controls; Firmicutes were less abundant and Fusobacteria and Proteobacteria were elevated in patients with INS, whereas butyrate-producing bacteria such as Lachnospira and Roseburia were more abundant in the healthy control group. In contrast, the bacterial groups Providencia and Myroides are more common in patients with INS [46]. The apparent difference in the relative abundance of butyric acid-producing bacteria suggests that this variation is not uncommon. A decrease in probiotics and beneficial microbial metabolites can cause a decrease in intestinal homeostasis. A decrease in probiotics and beneficial microbial metabolites can lead to a decline in intestinal homeostasis and even directly affect the differentiation and induction of immune cells [44]. Targeting probiotics and their products can provide new thinking for the identification and recurrence of INS.
Diabetic nephropathy
Diabetic nephropathy is one of the most severe and prognostic complications of diabetes mellitus. Disturbances in the gut microbiota have been observed in type 1 and type 2 diabetes, including significant reductions in the abundance of Lactobacillus and Bifidobacteria, which are involved in the maintenance of intestinal epithelial integrity. Other bacteria with high pathogenic potential, such as Clostridium and Bacteroidetes, were significantly increased in abundance [47]. In addition, intestinal flora that produces short-chain fatty acids (SCFAs) is significantly reduced in patients with diabetic nephropathy [48]. A meta-study suggested that H. pylori infection is associated with an increased risk of diabetic nephropathy and plays a role in the disease [49]. Moreover, antigens on the surface of Leptotrichia googfellowii have been found to stimulate CD8+ T cells to attack islets, which can promote the development of diabetic nephropathy. Short-chain fatty acid metabolites of Lactobacillus and Bifidobacterium can reduce insulin resistance and delay the progression of kidney disease [47]. Thus, the complex interactions between.
Renal aging and functional loss
Aging-related changes in the gut microbiome are mainly caused by systemic inflammation and aging of the immune system [50]. Microbiome alterations have also been demonstrated in the aging of the kidney. Gut microbes regulate local and systemic innate and adaptive immunity [51]. When the integrity of the gut barrier is breached, gut bacteria and other toxins can enter the body’s tissues and organs [52]. Immune cells and inflammatory factors produced during immune activation can contribute to the development of kidney disease [53]. Moreover, changes in the composition of the flora and metabolite production by the gut microbiota can promote inflammation, oxidative stress, and fibrosis in the kidneys. Dysregulation of the intestinal flora can lead to the production of uremic toxins such as indoxyl sulfate, p-cresol sulfate, and trimethylamine-N-oxide (TMAO). The toxin uremia can induce the production of pro-inflammatory factors that trigger inflammation and also promotes cellular aging and kidney fibrosis [54, 55]. TMAO is also considered to be a central link between the gut microbiome and kidney disease [56]. TMAO can promote the development of CKD by inducing inflammation and oxidative stress, upregulating scavenger receptors, and inhibiting reverse cholesterol transport [57]. In addition, reductions in intestinal bacteria that produce SCFAs, particularly butyric acid, have been observed in several kidney diseases. SCFAs are involved in the maintenance of the integrity of the intestinal barrier [58]. SCFAs can also attenuate the activation of NFĸB, inhibit the production of proinflammatory factors and regulate the activity of Tregs [59, 60]. The SCFA butyrate All Rights Reserved can also enhance mitochondrial activity, activate intestinal gluconeogenesis, and regulate epigenetic processes by inhibiting histone deacetylases. Thus, butyrate is considered to be a beneficial anti-aging metabolite [61]. A stable intestinal microbial structure is an important component of the gut microbiota. When this homeostasis is disrupted, the gut microbiota is responsible for the abnormal immune activation and inflammatory state in the body. The transformed role of gut microbes and their metabolites in these two distinct states also demonstrates the great potential of microbes in anti-aging research.
The microbiome — a new therapeutic target for renal injury and aging?
Microbiome research has broadened the diagnostic and treatment options for kidney injury (Figure 3). Evidence indicates the colon-kidney axis plays an important role in renal injury and imbalances in the intestinal flora are implicated in the pathophysiological process of kidney disease, which suggests that the restoration of bacterial homeostasis may be an effective treatment for kidney disease. Probiotics may represent an important potential treatment. Studies have shown that probiotics can effectively reduce the concentration of uremic toxins, especially p-cresol sulphate and p-indoxyl sulphate, in patients with CKD [62]. Moreover, probiotics can reduce the levels of inflammatory markers in the host and affect the immune system [63, 64]. Lactobacillus salivarius BP121 and Lactobacillus were shown to downregulate renal inflammatory mediators and reduce oxidative stress [65, 66]. Oral probiotics such as L. plantarum and L. brevis were also found to slow the progression of CKD and KS [67, 68]. Synbiotics are a combination of probiotics and prebiotics. When combined with low protein therapy, synbiotics could reduce the rate of progression of CKD, lead to significant enrichment of Bifidobacterium, reduce the abundance of Rumencoccus, and improve the microbial structure of the feces [69, 70]. SCFAs, the final metabolite of the fermentation of complex polysaccharides by the intestinal flora, also play an important role in renal function. SCFAs have strong anti-inflammatory properties and immunomodulatory effects [71, 72]. Supplementation with SCFAs has been shown to prevent the progression of AKI and subsequent CKD [73]. Fecal microbiome transplantation (FMT) is also considered to be an effective therapy to restore homeostasis to a disrupted microbiome. FMT treatment ameliorated intestinal microbiota disorder and limited the accumulation of uremic toxins in mouse models of CKD [74]. In the diabetic rat model, FMT also effectively reduced the levels of inflammatory factors and thereby attenuated inflammation and necrosis of the renal tubule interstitium in a model of diabetic nephropathy [75]. In addition, FMT also showed good therapeutic potential in patients with refractory IgA nephropathy [76, 77]. Overall, these microbiome-related therapeutic strategies have a high potential to reduce the incidence of kidney disease and improve patient outcomes, and may also represent new strategies to combat the effects of kidneyspecific aging.
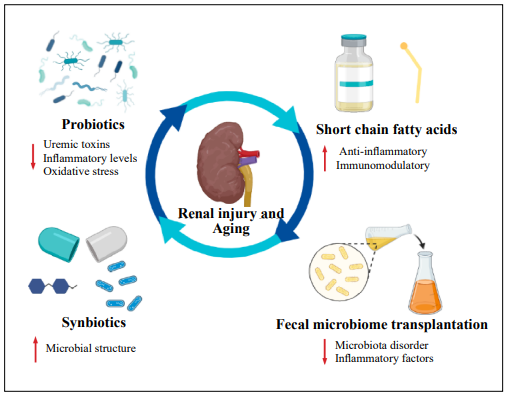
Figure 3. Potential therapeutic strategies for renal injury and aging associated with alterations to the microbiome and its metabolites.
Summary and perspectives
The microbiome is now viewed as a vital “organ” of the body that is closely related to human health and directly or indirectly affects the physiological functions of the body through multiple immune and metabolic pathways. Among the diseases associated with aging, kidney disease is not to be ignored. The decline in microbial diversity, decrease in probiotics and their metabolites, and increase in the relative abundance of disease-specific microorganisms are all signs of kidney aging and disease. The structure of the microbiome is related to health outcomes, and can more accurately describe abnormal states of the host than many traditional clinal markers. Detailed knowledge of microbial alterations may not only help to distinguish between diseased and non-diseased states but may also help to understand the response of the host to treatments and estimate prognosis. Further development of sequencing analysis technology may enable the disease course of individual patients to be more carefully defined and treatments to be selected more precisely. In addition, the microbiome itself has emerged as an important target of the disease. Many treatments targeting the microbiome have shown good efficacy in patients with nephrosis. A few microbial markers of renal dysfunction have been well explored. However, more advanced platforms to collate and analyze such markers and validation of the results in larger clinical cohorts are necessary to identify accurate microbial markers. These efforts may help to uncover the promising potential of microbial research to improve the diagnosis and treatment of kidney diseases and aging.
Declarations
Authors’ contributions
All authors contributed equally. All authors contributed to the manuscript and agreed to submit the manuscript.
Availability of data and materials
Not applicable.
Financial support and sponsorship
This work is supported by the National Key Research and Development Project of China (2018YFC2000500/03), the Natural Science Foundation of Shanghai (21ZR1409200), and the grants for a disciplined leader of Shanghai Municipal Health Commission National (2022XD051). Dr. C Zhao is supported by outstanding talent from Fudan University (2015).
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
References
1. Pflughoeft KJ, & Versalovic J. Human microbiome in health and disease. Annu Rev Pathol, 2012, 7: 99-122. [Crossref]
2. Sender R, Fuchs S, & Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell, 2016, 164(3): 337-340. [Crossref]
3. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature, 2014, 505(7484): 559-563. [Crossref]
4. Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, et al. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med Sci Sports Exerc, 2018, 50(4): 747-757. [Crossref]
5. Dethlefsen L, McFall-Ngai M, & Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature, 2007, 449(7164): 811-818. [Crossref]
6. Grice EA, & Segre JA. The skin microbiome. Nat Rev Microbiol, 2011, 9(4): 244-253. [Crossref]
7. Kort R, Caspers M, van de Graaf A, van Egmond W, Keijser B, & Roeselers G. Shaping the oral microbiota through intimate kissing. Microbiome, 2014, 2: 41. [Crossref]
8. Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol, 2011, 12(5): R50. [Crossref]
9. Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio, 2015, 6(5): e01578-01515. [Crossref]
10. Zheng Y, Fang Z, Xue Y, Zhang J, Zhu J, Gao R, et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes, 2020, 11(4): 1030-1042. [Crossref]
11. Liu H, Chen X, Hu X, Niu H, Tian R, Wang H, et al. Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome, 2019, 7(1): 68. [Crossref]
12. Yan Q, Gu Y, Li X, Yang W, Jia L, Chen C, et al. Alterations of the Gut Microbiome in Hypertension. Front Cell Infect Microbiol, 2017, 7: 381. [Crossref]
13. Cryan JF, O’Riordan KJ, Sandhu K, Peterson V, & Dinan TG. The gut microbiome in neurological disorders. Lancet Neurol, 2020, 19(2): 179-194. [Crossref]
14. Konturek PC, Haziri D, Brzozowski T, Hess T, Heyman S, Kwiecien S, et al. Emerging role of fecal microbiota therapy in the treatment of gastrointestinal and extragastrointestinal diseases. J Physiol Pharmacol, 2015, 66(4): 483-491.
15. Rehman T. Role of the gut microbiota in age-related chronic inflammation. Endocr Metab Immune Disord Drug Targets, 2012, 12(4): 361-367. [Crossref]
16. Xu R, Shang N, & Li P. In vitro and in vivo antioxidant activity of exopolysaccharide fractions from Bifidobacterium animalis RH. Anaerobe, 2011, 17(5): 226-231. [Crossref]
17. Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, et al. Gut Microbiota and Extreme Longevity. Curr Biol, 2016, 26(11): 1480-1485. [Crossref]
18. Glodny B, Unterholzner V, Taferner B, Hofmann KJ, Rehder P, Strasak A, et al. Normal kidney size and its influencing factors - a 64-slice MDCT study of 1.040 asymptomatic patients. BMC Urol, 2009, 9: 19. [Crossref]
19. Bao YW, Yuan Y, Chen JH, & Lin WQ. Kidney disease models: tools to identify mechanisms and potential therapeutic targets. Zool Res, 2018, 39(2): 72-86. [Crossref]
20. Ren Z, Fan Y, Li A, Shen Q, Wu J, Ren L, et al. Alterations of the Human Gut Microbiome in Chronic Kidney Disease. Adv Sci (Weinh), 2020, 7(20): 2001936. [Crossref]
21. Webster AC, Nagler EV, Morton RL, & Masson P. Chronic Kidney Disease. Lancet, 2017, 389(10075): 1238-1252. [Crossref]
22. Pasala S, & Carmody JB. How to use… serum creatinine, cystatin C and GFR. Arch Dis Child Educ Pract Ed, 2017, 102(1): 37-43. [Crossref]
23. Pichaiwong W, Homsuwan W, & Leelahavanichkul A. The prevalence of normoalbuminuria and renal impairment in type 2 diabetes mellitus . Clin Nephrol, 2019, 92(2): 73-80. [Crossref]
24. Jiang S, Xie S, Lv D, Wang P, He H, Zhang T, et al. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci Rep, 2017, 7(1): 2870. [Crossref]
25. Hu X, Ouyang S, Xie Y, Gong Z, & Du J. Characterizing the gut microbiota in patients with chronic kidney disease. Postgrad Med, 2020, 132(6): 495-505. [Crossref]
26. Hu Q, Wu K, Pan W, Zeng Y, Hu K, Chen D, et al. Intestinal flora alterations in patients with early chronic kidney disease: a case-control study among the Han population in southwestern China. J Int Med Res, 2020, 48(6): 300060520926033. [Crossref]
27. Wu IW, Gao SS, Chou HC, Yang HY, Chang LC, Kuo YL, et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics, 2020, 10(12): 5398-5411. [Crossref]
28. Wu IW, Lin CY, Chang LC, Lee CC, Chiu CY, Hsu HJ, et al. Gut Microbiota as Diagnostic Tools for Mirroring Disease Progression and Circulating Nephrotoxin Levels in Chronic Kidney Disease: Discovery and Validation Study. Int J Biol Sci, 2020, 16(3): 420-434. [Crossref]
29. Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, Murashita K, et al. Metagenomic profiling of gut microbiome in early chronic kidney disease. Nephrol Dial Transplant, 2021, 36(9): 1675-1684. [Crossref]
30. Lai KN, Tang SC, Schena FP, Novak J, Tomino Y, Fogo AB, et al. IgA nephropathy. Nat Rev Dis Primers, 2016, 2: 16001. [Crossref]
31. Wyatt RJ, & Julian BA. IgA nephropathy. N Engl J Med, 2013, 368(25): 2402-2414. [Crossref]
32. Huang C, Li X, Wu J, Zhang W, Sun S, Lin L, et al. The landscape and diagnostic potential of T and B cell repertoire in Immunoglobulin A Nephropathy. J Autoimmun, 2019, 97: 100-107. [Crossref]
33. D’Amico G. Natural history of idiopathic IgA nephropathy: role of clinical and histological prognostic factors. Am J Kidney Dis, 2000, 36(2): 227-237. [Crossref]
34. Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med, 2018, 10(439) [Crossref]
35. Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet, 2014, 46(11): 1187-1196. [Crossref]
36. Grosserichter-Wagener C, Radjabzadeh D, van der Weide H, Smit KN, Kraaij R, Hays JP, et al. Differences in Systemic IgA Reactivity and Circulating Th Subsets in Healthy Volunteers With Specific Microbiota Enterotypes. Front Immunol, 2019, 10: 341. [Crossref]
37. De Angelis M, Montemurno E, Piccolo M, Vannini L, Lauriero G, Maranzano V, et al. Microbiota and metabolome associated with immunoglobulin A nephropathy (IgAN). PLoS One, 2014, 9(6): e99006. [Crossref]
38. Hu X, Du J, Xie Y, Huang Q, Xiao Y, Chen J, et al. Fecal microbiota characteristics of Chinese patients with primary IgA nephropathy: a cross-sectional study. BMC Nephrol, 2020, 21(1): 97. [Crossref]
39. Zhong Z, Tan J, Tan L, Tang Y, Qiu Z, Pei G, et al. Modifications of gut microbiota are associated with the severity of IgA nephropathy in the Chinese population. Int Immunopharmacol, 2020, 89(Pt B): 107085. [Crossref]
40. Shah NB, Nigwekar SU, Kalim S, Lelouvier B, Servant F, Dalal M, et al. The Gut and Blood Microbiome in IgA Nephropathy and Healthy Controls. Kidney360, 2021, 2(8): 1261-1274. [Crossref]
41. Yamaguchi H, Goto S, Takahashi N, Tsuchida M, Watanabe H, Yamamoto S, et al. Aberrant mucosal immunoreaction to tonsillar microbiota in immunoglobulin A nephropathy. Nephrol Dial Transplant, 2021, 36(1): 75- 86. [Crossref]
42. Noone DG, Iijima K, & Parekh R. Idiopathic nephrotic syndrome in children. Lancet, 2018, 392(10141): 61-74. [Crossref]
43. Müller-Deile J, & Schiffer M. Podocyte directed therapy of nephrotic syndrome-can we bring the inside out? Pediatr Nephrol, 2016, 31(3): 393-405. [Crossref]
44. Tsuji S, Akagawa S, Akagawa Y, Yamaguchi T, Kino J, Yamanouchi S, et al. Idiopathic nephrotic syndrome in children: role of regulatory T cells and gut microbiota. Pediatr Res, 2021, 89(5): 1185-1191. [Crossref]
45. Tsuji S, Suruda C, Hashiyada M, Kimata T, Yamanouchi S, Kitao T, et al. Gut Microbiota Dysbiosis in Children with Relapsing Idiopathic Nephrotic Syndrome. Am J Nephrol, 2018, 47(3): 164-170. [Crossref]
46. Zhang J, Luo D, Lin Z, Zhou W, Rao J, Li Y, et al. Dysbiosis of gut microbiota in adult idiopathic membranous nephropathy with nephrotic syndrome. Microb Pathog, 2020, 147: 104359. [Crossref]
47. Mertowska P, Mertowski S, Wojnicka J, Korona-Głowniak I, Grywalska E, Błażewicz A, et al. A Link between Chronic Kidney Disease and Gut Microbiota in Immunological and Nutritional Aspects. Nutrients, 2021, 13(10) [Crossref]
48. Yu W, Shang J, Guo R, Zhang F, Zhang W, Zhang Y, et al. The gut microbiome in differential diagnosis of diabetic kidney disease and membranous nephropathy. Ren Fail, 2020, 42(1): 1100-1110. [Crossref]
49. Wang F, Liu J, & Lv Z. Association of Helicobacter pylori infection with diabetes mellitus and diabetic nephropathy: a meta-analysis of 39 studies involving more than 20,000 participants. Scand J Infect Dis, 2013, 45(12): 930-938. [Crossref]
50. Vaiserman AM, Koliada AK, & Marotta F. Gut microbiota: A player in aging and a target for anti-aging intervention. Ageing Res Rev, 2017, 35: 36-45. [Crossref]
51. Maynard CL, Elson CO, Hatton RD, & Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature, 2012, 489(7415): 231-241. [Crossref]
52. Bezirtzoglou E, & Stavropoulou E. Immunology and probiotic impact of the newborn and young children intestinal microflora. Anaerobe, 2011, 17(6): 369-374. [Crossref]
53. Imig JD, & Ryan MJ. Immune and inflammatory role in renal disease. Compr Physiol, 2013, 3(2): 957-976. [Crossref]
54. Motojima M, Hosokawa A, Yamato H, Muraki T, & Yoshioka T. Uremic toxins of organic anions up-regulate PAI- 1 expression by induction of NF-kappaB and free radical in proximal tubular cells. Kidney Int, 2003, 63(5): 1671- 1680. [Crossref]
55. Adijiang A, Shimizu H, Higuchi Y, Nishijima F, & Niwa T. Indoxyl sulfate reduces klotho expression and promotes senescence in the kidneys of hypertensive rats. J Ren Nutr, 2011, 21(1): 105-109. [Crossref]
56. Aron-Wisnewsky J, & Clément K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol, 2016, 12(3): 169-181. [Crossref]
57. Naghipour S, Cox AJ, Peart JN, Du Toit EF, & Headrick JP. Trimethylamine N-oxide: heart of the microbiota-CVD nexus? Nutr Res Rev, 2021, 34(1): 125-146. [Crossref]
58. Wang HB, Wang PY, Wang X, Wan YL, & Liu YC. Butyrate enhances intestinal epithelial barrier function via upregulation of tight junction protein Claudin-1 transcription. Dig Dis Sci, 2012, 57(12): 3126-3135. [Crossref]
59. Chang PV, Hao L, Offermanns S, & Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A, 2014, 111(6): 2247-2252. [Crossref]
60. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun, 2015, 6: 7320. [Crossref]
61. Vaiserman AM, & Pasyukova EG. Epigenetic drugs: a novel anti-aging strategy? Front Genet, 2012, 3: 224. [Crossref]
62. Turroni S, Magnani M, Kc P, Lesnik P, Vidal H, & Heer M. Gut Microbiome and Space Travelers’ Health: State of the Art and Possible Pro/Prebiotic Strategies for Long-Term Space Missions. Front Physiol, 2020, 11: 553929. [Crossref]
63. Tayebi Khosroshahi H, Vaziri ND, Abedi B, Asl BH, Ghojazadeh M, Jing W, et al. Effect of high amylose resistant starch (HAM-RS2) supplementation on biomarkers of inflammation and oxidative stress in hemodialysis patients: a randomized clinical trial. Hemodial Int, 2018, 22(4): 492-500. [Crossref]
64. Wang IK, Wu YY, Yang YF, Ting IW, Lin CC, Yen TH, et al. The effect of probiotics on serum levels of cytokine and endotoxin in peritoneal dialysis patients: a randomised, double-blind, placebo-controlled trial. Benef Microbes, 2015, 6(4): 423-430. [Crossref]
65. Lee TH, Park D, Kim YJ, Lee I, Kim S, Oh CT, et al. Lactobacillus salivarius BP121 prevents cisplatin-induced acute kidney injury by inhibition of uremic toxins such as indoxyl sulfate and p-cresol sulfate via alleviating dysbiosis. Int J Mol Med, 2020, 45(4): 1130-1140. [Crossref]
66. Lee YJ, Li KY, Wang PJ, Huang HW, & Chen MJ. Alleviating chronic kidney disease progression through modulating the critical genus of gut microbiota in a cisplatininduced Lanyu pig model. J Food Drug Anal, 2020, 28(1): 103-114. [Crossref]
67. Zhu H, Cao C, Wu Z, Zhang H, Sun Z, Wang M, et al. The probiotic L. casei Zhang slows the progression of acute and chronic kidney disease. Cell Metab, 2021, 33(10): 2091-2093. [Crossref]
68. Campieri C, Campieri M, Bertuzzi V, Swennen E, Matteuzzi D, Stefoni S, et al. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int, 2001, 60(3): 1097-1105. [Crossref]
69. Rossi M, Johnson DW, Morrison M, Pascoe EM, Coombes JS, Forbes JM, et al. Synbiotics Easing Renal Failure by Improving Gut Microbiology (SYNERGY): A Randomized Trial. Clin J Am Soc Nephrol, 2016, 11(2): 223-231. [Crossref]
70. Ebrahim Z, Proost S, Tito RY, Raes J, Glorieux G, Moosa MR, et al. The Effect of ß-Glucan Prebiotic on Kidney Function, Uremic Toxins and Gut Microbiome in Stage 3 to 5 Chronic Kidney Disease (CKD) Predialysis Participants: A Randomized Controlled Trial. Nutrients, 2022, 14(4) [Crossref]
71. Carney EF. Acute kidney injury. Protective role of gut microbial SCFAs. Nat Rev Nephrol, 2015, 11(3): 127. [Crossref]
72. Huang W, Zhou L, Guo H, Xu Y, & Xu Y. The role of shortchain fatty acids in kidney injury induced by gut-derived inflammatory response. Metabolism, 2017, 68: 20-30. [Crossref]
73. Liu Y, Li YJ, Loh YW, Singer J, Zhu W, Macia L, et al. Fiber Derived Microbial Metabolites Prevent Acute Kidney Injury Through G-Protein Coupled Receptors and HDAC Inhibition. Front Cell Dev Biol, 2021, 9: 648639. [Crossref]
74. Barba C, Soulage CO, Caggiano G, Glorieux G, Fouque D, & Koppe L. Effects of Fecal Microbiota Transplantation on Composition in Mice with CKD. Toxins (Basel), 2020, 12(12). [Crossref]
75. Hu ZB, Lu J, Chen PP, Lu CC, Zhang JX, Li XQ, et al. Dysbiosis of intestinal microbiota mediates tubulointerstitial injury in diabetic nephropathy via the disruption of cholesterol homeostasis. Theranostics, 2020, 10(6): 2803- 2816. [Crossref]
76. Zhao J, Bai M, Yang X, Wang Y, Li R, & Sun S. Alleviation of refractory IgA nephropathy by intensive fecal microbiota transplantation: the first case reports. Ren Fail, 2021, 43(1): 928-933. [Crossref]
77. Zhi W, Song W, Abdi Saed Y, Wang Y, & Li Y. Fecal Capsule as a Therapeutic Strategy in IgA Nephropathy: A Brief Report. Front Med (Lausanne), 2022, 9: 914250. [Crossref]
Supplementary
Table S1
Search strategies and results.
| Pubmed | Results |
|---|---|
| (Bacteroides) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 55 |
| (Faecalibacterium) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 15 |
| (Pseudomonas) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 340 |
| (Eubacterium) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 4 |
| (Parabacteroides) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 7 |
| (Alistipes) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 7 |
| (Ruminococcus) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 16 |
| (Bifidobacterium) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 41 |
| (Bacteroides) [All Fields] AND ((Immunoglobulin A nephropathy) [All Fields] OR (IgAN) [All Fields]) | 5 |
| (Faecalibacterium) [All Fields] AND ((Immunoglobulin A nephropathy) [All Fields] OR (IgAN) [All Fields]) | 0 |
| (Eubacterium) [All Fields] AND ((Immunoglobulin A nephropathy) [All Fields] OR (IgAN) [All Fields]) | 1 |
| (Parabacteroides) [All Fields] AND ((Immunoglobulin A nephropathy) [All Fields] OR (IgAN) [All Fields]) | 1 |
| (Alistipes) [All Fields] AND ((Immunoglobulin A nephropathy) [All Fields] OR (IgAN) [All Fields]) | 1 |
| (Ruminococcus) [All Fields] AND ((Immunoglobulin A nephropathy) [All Fields] OR (IgAN) [All Fields]) | 2 |
| (Bifidobacterium) [All Fields] AND ((Immunoglobulin A nephropathy) [All Fields] OR (IgAN) [All Fields]) | 3 |
| (Bacteroides) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 7 |
| (Faecalibacterium) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 0 |
| (Pseudomonas) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 26 |
| (Eubacterium) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 1 |
| (Parabacteroides) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 3 |
| (Alistipes) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 1 |
| (Ruminococcus) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 3 |
| (Bifidobacterium) [All Fields] AND ((Idiopathic nephrotic syndrome) [All Fields] OR (INS) [All Fields]) | 3 |
| (Bacteroides) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 13 |
| (Faecalibacterium) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 3 |
| (Pseudomonas) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 73 |
| (Eubacterium) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 4 |
| (Parabacteroides) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 0 |
| (Alistipes) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 0 |
| (Ruminococcus) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 2 |
| (Bifidobacterium) [All Fields] AND ((kidney stone disease) [All Fields] OR (Kidney stones) [All Fields] OR (KS) [All Fields]) | 19 |
| (Bacteroides) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 8 |
| (Faecalibacterium) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 2 |
| (Pseudomonas) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 219 |
| (Eubacterium) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 1 |
| (Parabacteroides) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 1 |
| (Alistipes) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 0 |
| (Ruminococcus) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 1 |
| (Bifidobacterium) [All Fields] AND ((Acute kidney injury) [All Fields] OR(AKI) [All Fields]) | 9 |
| (Bacteroides) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic kidney diseases) [All Fields]) | 39 |
| (Faecalibacterium) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic kidney diseases) [All Fields]) | 5 |
| (Pseudomonas) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic kidney diseases) [All Fields]) | 103 |
| (Eubacterium) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic kidney diseases) [All Fields]) | 7 |
| (Bacteroides) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 55 |
| (Faecalibacterium) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 15 |
| (Pseudomonas) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 340 |
| (Eubacterium) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 4 |
| (Parabacteroides) [All Fields] AND ((CKD) [All Fields] OR (Chronic kidney disease) [All Fields]) | 7 |
| (Parabacteroides) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic kidney diseases) [All Fields]) | 2 |
| (Alistipes) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic kidney diseases) [All Fields]) | 2 |
| (Ruminococcus) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic | 3 |
| (Bifidobacterium) [All Fields] AND ((Diabetic nephropathy) [All Fields] OR (DN) [All Fields] OR (DKD) [All Fields] OR (diabetic kidney diseases) [All Fields]) | 66 |
Table S2
Comparison of gut bacteria between patients with kidney diseases and healthy controls at the genus level.
| Genus | Significant higher in CKD | Significant lower or no difference in CKD | Significant higher in IgAN | Significant lower or no difference in IgAN | ||||||||||||
| n | Citation | n | Citation | n | Citation | n | Citation | |||||||||
| Bacteroides | 8 | [1-8] | 4 | [9-12] | 2 | [13, 14] | 2 | [15, 16] | ||||||||
| Faecalibacterium | 1 | [17] | 8 | [3, 4, 12, 18-21] | 1 | [15] | / | / | ||||||||
| Pseudomonas | 1 | [22] | / | / | / | / | / | / | ||||||||
| Eubacterium | 2 | [7, 17] | / | / | / | / | 1 | [13] | ||||||||
| Parabacteroides | 2 | [6, 23] | / | / | 1 | [13] | / | / | ||||||||
| Alistipes | 2 | [2, 24] | 1 | [12] | / | / | 2 | [13, 25] | ||||||||
| Ruminococcus | 7 | [2, 6, 7, 12, 17, 24, 26] | 1 | [1] | 1 | [13] | 1 | [25] | ||||||||
| Bifidobacterium | 4 | [8, 12, 21, 27] | 1 | [28] | / | / | 3 | [13, 14, 16] | ||||||||
| Genus | Significant higher in INS | Significant lower or no difference in INS | Significant higher in KS | Significant lower or no difference in KS | ||||||||||||
| n | Citation | n | Citation | n | Citation | n | Citation | |||||||||
| Bacteroides | / | / | / | / | 4 | [29-32] | / | / | ||||||||
| Faecalibacterium | / | / | / | / | 1 | [31] | 4 | [29, 32-34] | ||||||||
| Pseudomonas | / | / | / | / | 1 | [35] | / | / | ||||||||
| Eubacterium | 1 | [36] | 2 | [37, 38] | 1 | [34] | 1 | [31] | ||||||||
| Parabacteroides | 2 | [26, 36] | / | / | / | / | / | / | ||||||||
| Alistipes | / | / | 1 | [38] | / | / | 1 | [32] | ||||||||
| Ruminococcus | / | / | / | / | / | / | / | / | ||||||||
| Bifidobacterium | / | / | / | / | / | / | 3 | [29, 30, 34] | ||||||||
| Genus | Significant higher in AKL | Significant lower or no difference in AKI | Significant higher in DN | Significant lower or no difference in DN | ||||||||||||
| n | Citation | n | Citation | n | Citation | n | Citation | |||||||||
| Bacteroides | / | / | / | / | 2 | [39, 40] | 1 | [41] | ||||||||
| Faecalibacterium | / | / | / | / | / | / | 3 | [39, 41, 42] | ||||||||
| Pseudomonas | / | / | / | / | / | / | / | / | ||||||||
| Eubacterium | / | / | / | / | / | / | 1 | [39] | ||||||||
| Parabacteroides | / | / | / | / | / | / | / | / | ||||||||
| Alistipes | / | / | / | / | 1 | [39] | / | / | ||||||||
| Ruminococcus | / | / | / | / | 2 | [41, 42] | / | / | ||||||||
| Bifidobacterium | / | / | / | / | 2 | [39, 42] | / | / | ||||||||
References
1. Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, Murashita K, et al. Metagenomic profiling of gut
microbiome in early chronic kidney disease. Nephrol Dial Transplant, 2021, 36(9):1675-1684. [Crossref]
2. Wu IW, Lin CY, Chang LC, Lee CC, Chiu CY, Hsu HJ, et al. Gut Microbiota as Diagnostic Tools for Mirroring
Disease Progression and Circulating Nephrotoxin Levels in Chronic Kidney Disease: Discovery and Validation Study.Int J Biol Sci, 2020, 16(3):420-434. [Crossref]
3. Hu J, Zhong X, Yan J, Zhou D, Qin D, Xiao X, et al. High-throughput sequencing analysis of intestinal flora changes
in ESRD and CKD patients. BMC Nephrol, 2020, 21(1):12. [Crossref]
4. Jiang S, Xie S, Lv D, Wang P, He H, Zhang T, et al. Alteration of the gut microbiota in Chinese population with
chronic kidney disease. Sci Rep, 2017, 7(1):2870. [Crossref]
5. Shivani S, Kao CY, Chattopadhyay A, Chen JW, Lai LC, Lin WH, et al. Uremic Toxin-Producing Bacteroides Species
Prevail in the Gut Microbiota of Taiwanese CKD Patients: An Analysis Using the New Taiwan Microbiome Baseline.
Front Cell Infect Microbiol, 2022, 12:726256. [Crossref]
6. Lun H, Yang W, Zhao S, Jiang M, Xu M, Liu F, et al. Altered gut microbiota and microbial biomarkers associated
with chronic kidney disease. Microbiologyopen, 2019, 8(4):e00678. [Crossref]
7. Hu Q, Wu K, Pan W, Zeng Y, Hu K, Chen D, et al. Intestinal flora alterations in patients with early chronic
kidney disease: a case-control study among the Han population in southwestern China. J Int Med Res, 2020,
48(6):300060520926033. [Crossref]
8. Gryp T, Huys GRB, Joossens M, Van Biesen W, Glorieux G, & Vaneechoutte M. Isolation and Quantification
of Uremic Toxin Precursor-Generating Gut Bacteria in Chronic Kidney Disease Patients. International journal of
molecular sciences, 2020, 21(6). [Crossref]
9. Wu IW, Gao SS, Chou HC, Yang HY, Chang LC, Kuo YL, et al. Integrative metagenomic and metabolomic analyses
reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics, 2020, 10(12):5398-5411.
[Crossref]
10. Lohia S, Vlahou A, & Zoidakis J. Microbiome in Chronic Kidney Disease (CKD): An Omics Perspective. Toxins
(Basel), 2022, 14(3). [Crossref]
11. Jiang S, Wang B, Sha T, & Li X. Changes in the Intestinal Microbiota in Patients with Stage 5 Chronic Kidney
Disease on a Low-Protein Diet and the Effects of Human to Rat Fecal Microbiota Transplantation. Med Sci Monit, 2020,
26:e921557. [Crossref]
12. Liu F, Xu X, Chao L, Chen K, Shao A, Sun D, et al. Alteration of the Gut Microbiome in Chronic Kidney Disease
Patients and Its Association With Serum Free Immunoglobulin Light Chains. Front Immunol, 2021, 12:609700. [Crossref]
13. Shah NB, Nigwekar SU, Kalim S, Lelouvier B, Servant F, Dalal M, et al. The Gut and Blood Microbiome in IgA
Nephropathy and Healthy Controls. Kidney 360, 2021, 2(8):1261-1274. [Crossref]
14. Zhong Z, Tan J, Tan L, Tang Y, Qiu Z, Pei G, et al. Modifications of gut microbiota are associated with the severity
of IgA nephropathy in the Chinese population. Int Immunopharmacol, 2020, 89(Pt B):107085.[Crossref]
15. Wu H, Tang D, Zheng F, Li S, Zhang X, Yin L, et al. Identification of a novel interplay between intestinal bacteria
and metabolites in Chinese patients with IgA nephropathy via integrated microbiome and metabolome approaches. Ann
Transl Med, 2021, 9(1):32. [Crossref]
16. De Angelis M, Montemurno E, Piccolo M, Vannini L, Lauriero G, Maranzano V, et al. Microbiota and metabolome
associated with immunoglobulin A nephropathy (IgAN). PloS one, 2014, 9(6):e99006. [Crossref]
17. Margiotta E, Miragoli F, Callegari ML, Vettoretti S, Caldiroli L, Meneghini M, et al. Gut microbiota composition
and frailty in elderly patients with Chronic Kidney Disease. PloS one, 2020, 15(4):e0228530. [Crossref]
18. Guirong YE, Minjie Z, Lixin YU, Junsheng YE, Lin Y, & Lisha S. [Gut microbiota in renal transplant recipients,
patients with chronic kidney disease and healthy subjects]. Nan Fang Yi Ke Da Xue Xue Bao, 2018, 38(12):1401-1408.
19. Jiang S, Xie S, Lv D, Zhang Y, Deng J, Zeng L, et al. A reduction in the butyrate producing species Roseburia spp.
and Faecalibacterium prausnitzii is associated with chronic kidney disease progression. Antonie Van Leeuwenhoek,
2016, 109(10):1389-1396. [Crossref]
20. Zhao J, Ning X, Liu B, Dong R, Bai M, & Sun S. Specific alterations in gut microbiota in patients with chronic
kidney disease: an updated systematic review. Ren Fail, 2021, 43(1):102-112. [Crossref]
21. Li Y, Su X, Zhang L, Liu Y, Shi M, Lv C, et al. Dysbiosis of the gut microbiome is associated with CKD5 and
correlated with clinical indices of the disease: a case-controlled study. J Transl Med, 2019, 17(1):228. [Crossref]
22. Wang F, Jiang H, Shi K, Ren Y, Zhang P, & Cheng S. Gut bacterial translocation is associated with
microinflammation in end-stage renal disease patients. Nephrology (Carlton), 2012, 17(8):733-738. [Crossref]
23. Hu X, Ouyang S, Xie Y, Gong Z, & Du J. Characterizing the gut microbiota in patients with chronic kidney disease.
Postgrad Med, 2020, 132(6):495-505. [Crossref]
24. Wang X, Yang S, Li S, Zhao L, Hao Y, Qin J, et al. Aberrant gut microbiota alters host metabolome and impacts
renal failure in humans and rodents. Gut, 2020, 69(12):2131-2142. [Crossref]
25. Chai L, Luo Q, Cai K, Wang K, & Xu B. Reduced fecal short-chain fatty acids levels and the relationship with gut
microbiota in IgA nephropathy. BMC Nephrol, 2021, 22(1):209. [Crossref]
26. Zhang J, Luo D, Lin Z, Zhou W, Rao J, Li Y, et al. Dysbiosis of gut microbiota in adult idiopathic membranous
nephropathy with nephrotic syndrome. Microb Pathog, 2020, 147:104359. [Crossref]
27. Lin TY, & Hung SC. Association of subjective global assessment of nutritional status with gut microbiota in
hemodialysis patients: a case-control study. Nephrol Dial Transplant, 2021, 36(6):1104-1111. [Crossref]
28. Hanifi GR, Samadi Kafil H, Tayebi Khosroshahi H, Shapouri R, & Asgharzadeh M. Bifidobacteriaceae Family Diversity in Gut Microbiota of Patients with Renal Failure. Arch Razi Inst, 2021, 76(3):521-528. [Crossref]
29. Stanford J, Charlton K, Stefoska-Needham A, Ibrahim R, & Lambert K. The gut microbiota profile of adults with
kidney disease and kidney stones: a systematic review of the literature. BMC Nephrol, 2020, 21(1):215. [Crossref]
30. Zhao E, Zhang W, Geng B, You B, Wang W, & Li X. Intestinal dysbacteriosis leads to kidney stone disease. Mol Med
Rep, 2021, 23(3). [Crossref]
31. Stern JM, Moazami S, Qiu Y, Kurland I, Chen Z, Agalliu I, et al. Evidence for a distinct gut microbiome in kidney
stone formers compared to non-stone formers. Urolithiasis, 2016, 44(5):399-407. [Crossref]
32. Chen F, Bao X, Liu S, Ye K, Xiang S, Yu L, et al. Gut microbiota affect the formation of calcium oxalate renal
calculi caused by high daily tea consumption. Appl Microbiol Biotechnol, 2021, 105(2):789-802. [Crossref]
33. Ticinesi A, Milani C, Guerra A, Allegri F, Lauretani F, Nouvenne A, et al. Understanding the gut-kidney axis
in nephrolithiasis: an analysis of the gut microbiota composition and functionality of stone formers. Gut, 2018,
67(12):2097-2106. [Crossref]
34. Kim HN, Kim JH, Chang Y, Yang D, Joo KJ, Cho YS, et al. Gut microbiota and the prevalence and incidence of
renal stones. Scientific reports, 2022, 12(1):3732. [Crossref]
35. Yuan C, Jin X, He Y, Liu Y, Xiang L, & Wang K. Association of dietary patterns with gut microbiota in kidney stone
and non-kidney stone individuals. Urolithiasis, 2022, 50(4):389-399. [Crossref]
36. He H, Lin M, You L, Chen T, Liang Z, Li D, et al. Gut Microbiota Profile in Adult Patients with Idiopathic
Nephrotic Syndrome. Biomed Res Int, 2021, 2021:8854969. [Crossref]
37. Tsuji S, Suruda C, Hashiyada M, Kimata T, Yamanouchi S, Kitao T, et al. Gut Microbiota Dysbiosis in Children
with Relapsing Idiopathic Nephrotic Syndrome. Am J Nephrol, 2018, 47(3):164-170. [Crossref]
38. Tsuji S, Akagawa S, Akagawa Y, Yamaguchi T, Kino J, Yamanouchi S, et al. Idiopathic nephrotic syndrome in
children: role of regulatory T cells and gut microbiota. Pediatr Res, 2021, 89(5):1185-1191. [Crossref]
39. Zhang L, Wang Z, Zhang X, Zhao L, Chu J, Li H, et al. Alterations of the Gut Microbiota in Patients with Diabetic
Nephropathy. Microbiol Spectr, 2022:e0032422. [Crossref]
40. He X, Sun J, Liu C, Yu X, Li H, Zhang W, et al. Compositional Alterations of Gut Microbiota in Patients with
Diabetic Kidney Disease and Type 2 Diabetes Mellitus. Diabetes Metab Syndr Obes, 2022, 15:755-765. [Crossref]
41. Wang Y, Ye X, Ding D, & Lu Y. Characteristics of the intestinal flora in patients with peripheral neuropathy
associated with type 2 diabetes. J Int Med Res, 2020, 48(9):300060520936806. [Crossref]
42. Chen W, Zhang M, Guo Y, Wang Z, Liu Q, Yan R, et al. The Profile and Function of Gut Microbiota in Diabetic
Nephropathy. Diabetes Metab Syndr Obes, 2021, 14:4283-4296. [Crossref]