Open Access | Review
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
The role of microRNAs in the occurrence and development of esophageal squamous cell carcinoma
*Corresponding author: Yan-ru Qin, MD. Mailing address: Department of Oncology, The First Affiliated Hospital of Zhengzhou University, 1 Jianshe East Road, Zhengzhou, Henan, P.R. China
E-mail:Yanruqin@163.com
Received: 20 October 2017 Accepted: 25 December 2017
DOI: 10.31491/CSRC.2017.12.001
Abstract
Esophageal carcinoma (EC) is a malignancy with high metastatic potential. Although the treatment has improved over the past few decades, the survival rate is still low, and there is a pressing need for targeted therapies. A variety of microRNA (miRNA) expression profiles and candidate genes have been studied in EC, which obviously supports the key role of miRNA in the pathogenesis of EC. In order to better understand the importance of miRNA in the development of EC, this article reviews the current information about many important miRNAs as they relate to the progression of EC.
Keywords
miRNA; esophageal carcinoma cell carcinoma; prognosis; chemosensitivity
Introduction
Esophageal carcinoma (EC) is a common cancer that causes a high rate of cancer-related mortality around the world. The five-year survival rate of patients with advanced stage or metastatic EC is reported to be below 20% [1]. EC is a malignant lesion formed by abnormal hyperplasia of the squamous epithelium or glandular epithelium of the esophagus, so EC usually occurs as either esophageal adenocarcinoma (EAC) or esophageal squamous cell carcinoma (ESCC) [2]. ESCC forms in squamous cells and occurs mainly in the upper 1/3 of the esophagus. However, EAC begins in glandular cells and usually forms in the lower part of the esophagus near the stomach. Of the two histological types of EC, ESCC is the more aggressive malignant tumor [3]. The incidence of EAC is rare in Asian and African countries where ESCC remains the dominant form. In China, more than 90% of the ECs are ESCC [4]. Although multimodulated treatment for ESCC patients exists, such as surgery, chemotherapy, and radiation therapy, the prognosis is still poor. Therefore, this review aims to enhance our understanding of the molecular mechanisms associated with the development and progression of ESCC.
MicroRNAs (miRNAs) are endogenous small noncoding RNAs which exist in various plants and animals [5]. They regulate gene expression by repressing the translation of target cellular transcripts, and they therefore take part in physiological and pathological processes that include cell proliferation, differentiation, and apoptosis. In the last century, scholars mainly paid attention on the protein-coding genes, and the existence of miRNAs was unknown. However, the small noncoding RNA lin-4, which was discovered in 1993, completely changed the opinion and perspective of scholars [6]. The biogenesis processing of these miRNAs happens in the nucleus/cytoplasm, while the final maturation occurs in the cytoplasm. Existing research has determined that miRNA genes are processed via both a regulated and a non-regulated biogenesis pathway and transcribed by RNA polymerase II. In the biological regulating processes, primary miRNAs (pri-miRNAs) are processed into a nucleus to generate approximately 70 nucleotide precursor miRNA (pre-miRNA) products by the ribonuclease III (RNase III) enzyme Drosha. While in the non-regulated biogenesis pathway, pre-miRNAs are instead generated by the mRNA splicing machinery, thereby avoiding Drosha digestion [7]. Currently, because the miRNA situation is very complicated due to the increased number of miRNA genes, it remains difficult to define their function clearly.
The changed expression of miRNA will regulate the oncogenes or the tumor suppressor expression, which will further influence cancer cell proliferation, apoptosis, migration, and invasion, among other things. This is the case with ESCC, in which there are various miRNA expression profiles that have been reported as possible useful biomarkers. Therefore, in this review, we summarized the characterization of miRNAs involved in ESCC, and we discussed the various miRNAs that could be biomarkers or therapeutic targets.
The role Of MiRNAs in the occurrence Of EC
A number of dysregulated miRNAs have been identified in ESCC, including the upregulation of miR-10b, miR- 151, miR-93, miR-21, miR-192, miR-194, and miR-106- 25 polycistron, and the downregulation of let-7c, miR- 125b, miR-27b, miR-145, miR-203, miR-100, miR-205, and miR-375. The majority of these are also abnormally expressed in other forms of cancer, and their target genes are widely studied in different cancers [8]. These abnormal miRNAs strongly suggest their important roles in ESCC development.
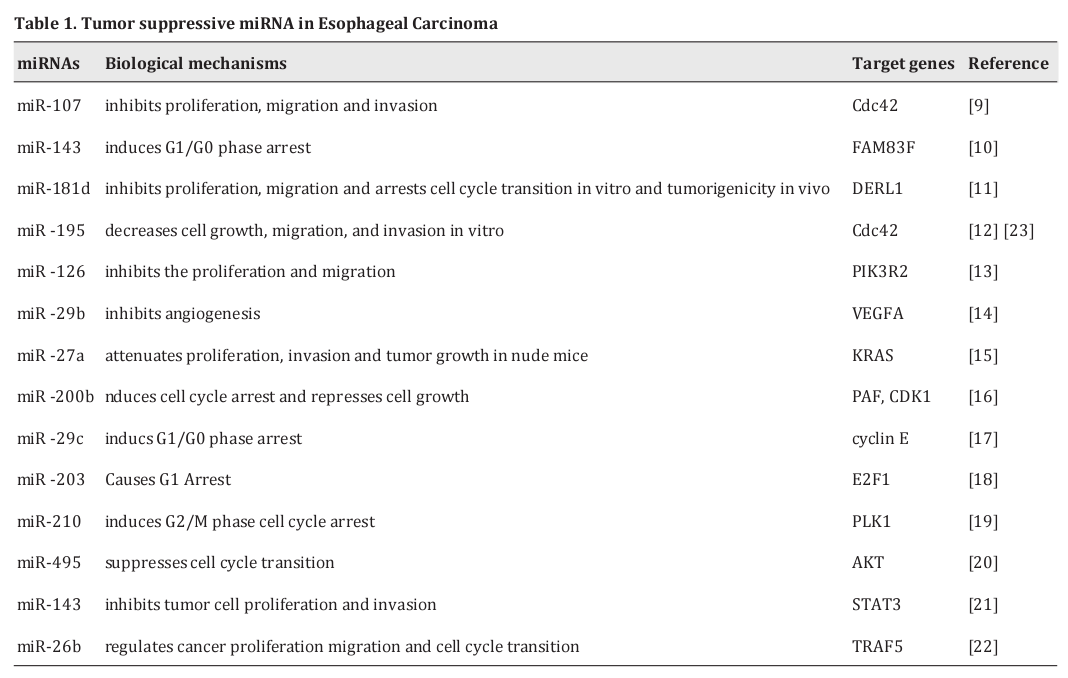
MiRNAs as tumor suppressors of EC
The tumor suppressive miRNA in EC is shown in Table 1. It has been determined that miR-107 functions as a tumor suppressor and inhibits the proliferation, migration, and invasion of ESCC cells [9]. MiR-143 plays the role of inhibiting tumors through inducing G1/ G0 phase arrest of ESCC cells by negatively regulating FAM83F expression [10]. MiR-181d is aberrantly downregulated in ESCC cell lines and performs a tumor suppressive function by inversely regulating its downstream target gene of DERL1 [11]. MiR-195 can target Cdc42 and serve as a tumor suppressor in ESCC [12]. MiR-126 is significantly downregulated in ESCC tissues compared to adjacent normal tissues, and the PI3K/AKT pathway has been found to be activated in ESCC tissues. Nie et al. [13] demonstrated that the PIK3R2 is a direct target of miR-126, which reveals that targeting miR-126 may be an effective strategy for ESCC diagnosis and treatment. VEGFA is upstream of the MAPK/ERK and PI3K/Akt signaling pathways, and miR-29b has been found to negatively modulate the MAPK/ERK and PI3K/Akt signaling pathways to inhibit angiogenesis in EC by targeting VEGFA [14]. MiR- 27a functions as a tumor suppressor by targeting KRAS, which further inhibits the KRAS-related ERK pathways [15]. MiR-200b has been reported to modulate Wnt/β- Catenin signaling by targeting PAF (PCNA-associated factor) in ESCC [16]. MiR-29c suppresses tumor growth by modulating cyclin E expression to induce cell cycle G(1)/G(0) arrest [17]. Zhang et al. [18] find that miR-203 is a target of E2F1 and regulated cell cycle arrest by taking part in a feedback loop with E2F1. MiR-210 has been reported to inhibit ESCC cell proliferation by targeting PLK1 to induce G2/M phase cell cycle arrest [19]. MiR- 495 suppresses cell cycle transition through targeting Akt1 in ESCC cells [20]. MiR-143 represses the cell cycle transition by targeting STAT3 in ESCC cells [21]. MiR-26b regulates cancer cell cycle transition by suppressing TRAF5 in ESCC cells [22].
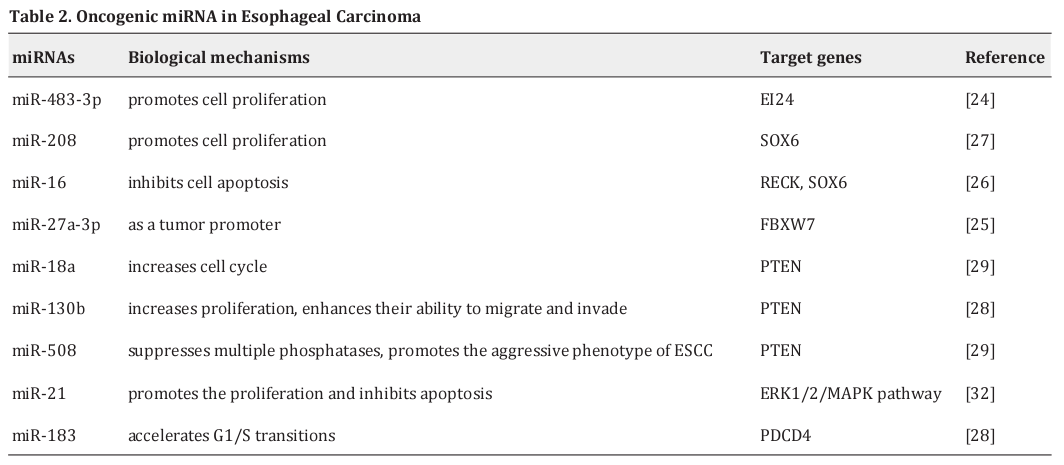
MiRNAs as oncogenes of EC
The oncogenic miRNA in EC is shown in Table 2. MiR-483-3p is upregulated in ESCC cell lines, and it promotes ESCC cell proliferation by directly targeting EI24, which suggests a potential strategy for miRNA-based ESCC therapy [24]. Wu et al. [25] discovered that miR-27a-3p, a tumor promoter which targets FBXW7, has a significantly inhibitory effect on the proliferation of Eca109 cells, indicating a potential therapeutic strategy for ESCC therapy. MiR-16 can inhibit ESCC cell apoptosis while promoting growth by the two genes, RECK and SOX6, which also play important roles in the etiopathogenesis of ESCC [26]. In ESCC cell lines and tissues, there is increased miR-208, which could promote ESCC cell proliferation. Further investigation found that miR-208 participates in ESCC carcinogenesis by targeting SOX6 [27]. MiR-183 accelerates G1/S transitions through inhibiting PDCD4 expression [28]. Phosphatase and tensin homolog (PTEN) is a tumor suppressor gene. Loss of its function is the most frequent genetic alteration in EC. The PI3KAkt-mTOR pathway regulates cellular growth, and PTEN is a negative regulator of this pathway. Zhang et al. [29] reported that miR-18a-increased cyclin D1 expression is associated with decreased PTEN, which has been demonstrated to be a direct target of miR-18a, indicating that upregulation of miR-18a promotes cell proliferation via regulating the PI3KAKT-mTOR signaling axis by targeting PTEN. MiR- 130b and miR-508 were also found to regulate p-Akt expression by targeting PTEN, thereby activating PI3K-Akt signaling [30,31]. In addition, miR-21 can activate the ERK1/2/MAPK pathway to promote Eca109 cell proliferation [32].
MicroRNAs and metastasis of EC
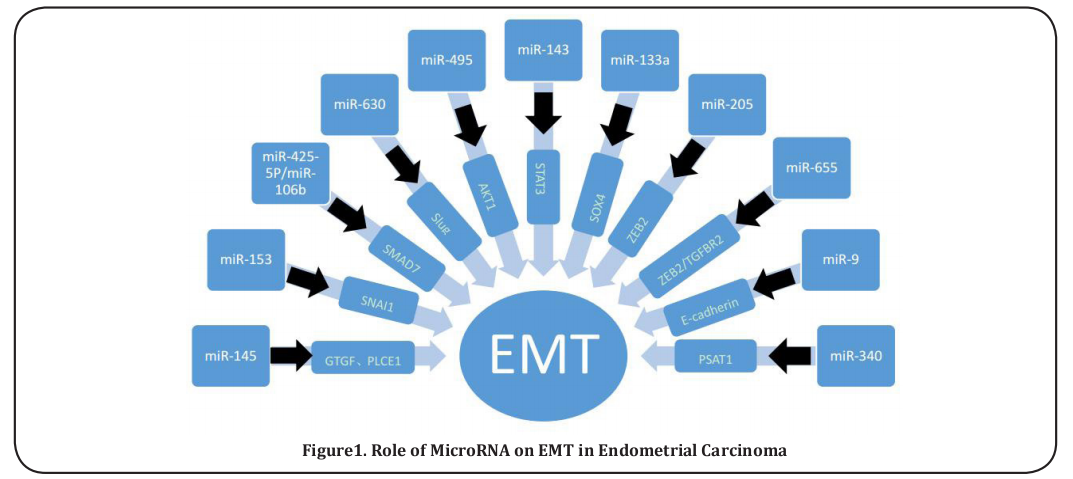
Epithelial-mesenchymal transition (EMT) is a signal pathway, which includes successive cell detaching, migrating, invading, dispersing, and residing, and has been classified as a hallmark of tumor metastasis. EMT is a crucial component of the invasive and metastatic properties of malignant tumor cells during tumor progression. One of the important mediators of EMT is the Snail1 protein (encoded by SNAI1), which facilitates the transition to the mesenchymal state by transcriptionally repressing the epithelial cell marker E-cadherin (Figure 1). It was reported that miR-153 can inhibit tumor progression in ESCC cells by targeting SNAI1 [33]. MiR-9 induces EMT in ESCC metastasis by targeting E-cadherin [34]. Slug is another EMT transcription factor, and miR-630 inhibits EMT by targeting Slug in ESCC [35]. Zeb2 (Sip1/Zfhx1b) is a member of the zinc-finger E-box-binding (ZEB) family of transcriptional repressors previously demonstrated to regulate EMT processes during embryogenesis and tumor progression. MiR-205 is an ESCC-specific miR that exerts tumor-suppressive activities with EMT inhibition by targeting ZEB2 [36]. MiR-655 is an EMT-suppressive miRNA targeting ZEB1 and TGFBR2 [37]. SOX4 knockdown delays TGF-β-induced EMT, and MiR- 133a suppresses the migration and invasion of EC cells by targeting the EMT regulator SOX4 [38].
Numerous signaling pathways are involved in the process of EMT in cancer, such as the SMAD7 pathway, STAT3 signaling pathway, PTEN/PI3K pathway, etc., which have a certain relationship with EMT. MiR-424-5p mediates EMT in ESCC via the SMAD7 pathway [39]. MiR-106b enhances the EMT process via downregulation of Smad7 in ESCC metastases [40]. The STAT3 signaling pathway plays an important role in the process of EMT, and miR-143 represses EMT signaling pathways by targeting STAT3 [21] in ESCC cells. Akt1 has been shown to inhibit EMT as well as cell motility, and miR-495 suppresses the EMT signaling pathway through targeting Akt1 in ESCC cells [20]. In addition, it was reported that miR-145 inhibits the EMT process of ESCC cells through the targeted regulation of CTGF and PLCE1 expression [41,42]. MiR-340 mediates inhibition of EMT in ESCC cells by modulating the expression of PSAT1 [43].
MicroRNAs as biomarkers in the diagnosis and treatment of EC
MiRNAs and the prognosis of EC
Considerable research has assessed miRNAs as prognostic factors in EC. A recent review summarized by Mei et al. [44] reported that prognostic miRNAs pair off with their target genes, which highlights the opportunities and challenges for miRNAs in the molecular diagnosis and target therapy of ESCC, such as the oncogenic miRNAs (miR-21 and miR-17-92 clusters) and the tumor suppressor miRNAs (miR-375, miR-133a, and miR-133b).
Over the past five years, many studies have identified some miRNAs that may be used as prognostic markers in clinics. There are 44 miRNAs, which Wang and his coworkers [45] revealed as being associated with the prognosis for ESCC. Among them, miR-143-3p and miR-145-5p were increased, while miR-182-5p and miR-455-5p were decreased, and all of them were demonstrated to be related to prognosis. There is an obviously downregulated miR-15a expression in ESCC tissues, and the low miR-15a expression has a significant correlation to shorter overall survival rates of ESCC patients [46]. Further, the multivariate analysis demonstrated that miR-15a is an independent prognostic factor for both overall survival (OS) and disease free survival (DFS). The low miR-7 expression in ESCC is an independent prognostic factor for poor survival, and miR-7 might be a therapeutic target in ESCC [47]. MiR-377 is located in chromosome region 14q32, which is frequently deleted in ESCC. A recent study by Li and his colleagues [48] reported that miR- 377 plays a key role in suppressing tumor initiation and progression, and their study confirmed miR-377 may represent a prognostic biomarker and therapeutic strategy for ESCC patients. Okumura et al. [49], using microarray analysis, identified miR-574-3p, miR- 106b, miR-1303, miR-1203, miR-1909, miR-204, miR-371-3p, and miR-886-3p in training set patients, which were differentially expressed in the patients with postoperative tumor relapse or those without it. Among these miRNAs, the upregulated miR-574- 3p showed the most significant association with non-relapse, which suggests that miR-574-3p is a predictor of postoperative outcome in ESCC patients. MiR-146a is significantly reduced in cancerous tissue and serum samples of ESCC patients, and miR-146a could be used as an independent prognostic factor for overall survival and progression-free survival [50]. Previous studies confirmed that miR-195 may act as a tumor suppressor in ESCC by regulating Cdc42 expression [12]. Also, further research suggests that aberrant miR- 195 and Cdc42 expression can serve as a promising unfavorable prognostic biomarker in ESCC [23]. The downregulated miR-100 in ESCC plays a crucial role in progression and prognosis of ESCC [51]. The members of the miR-17-92 cluster have been reported to be highly expressed in several cancers. MiR-18a, miR-19a, and miR-17 are members of the miR-17-92 cluster, and the univariate and multivariate analysis demonstrated that they may been prognostic indicators for progressionfree survival and overall survival, which indicates that miR-17, miR-18a, and miR-19a can serve as potential unfavorable prognostic biomarkers in ESCCs which are associated with some clinicopathologic factors [52]. In addition, the miR-18a that is located in the miR-17-92 cluster was further reported as being highly expressed in tissues of ESCC [53]. The miR-181b, miR-21, and miR- 146b expression in tumor tissue are associated with overall survival of patients, indicating that they may be used as prognostic classifiers for ESCC [54]. Measuring the levels of miRNAs in 64 EC patients who underwent neoadjuvant chemotherapy, Tanaka et al. concluded that the miR-200c expression in serum can predict the response to chemotherapy and the prognosis of EC patients who receive neoadjuvant chemotherapy [55]. Combining the expressions of miR-133a and miR- 133b were found to be an effective prognostic marker of ESCC [56].
Several recent studies demonstrated that miRNAs are stably detectable in plasma/serum. Detection of circulating miRNAs might provide new complementary tumor markers for ESCC. Circulating miR-155 in plasma may serve as a reliable, novel, and noninvasive biomarker for the early diagnosis and detection of EC [57]. The miR-18a that is highly expressed in tissues of ESCC could serve as a plasma biomarker in patients with ESCC [53]. Komatsu et al. [58] selected three oncogenic miRNAs (miR-21, miR-184, and miR-221) and one tumor suppressive miRNA (miR-375), which are frequently reported in ESCC, as candidate targets for a plasma miRNA assay. The results showed that patients with a high plasma level of miR-21 tend to have greater vascular invasion and that this indicates a high correlation with recurrence. Moreover, Komatsu and his colleagues [59] once again examined the association between plasma miRNA concentrations and prognosis retrospectively in 50 preoperative plasma samples that were collected from consecutive patients with ESCC who underwent curative esophagectomy. Their research concluded that circulating miR-21 and miR-375 could be reliable prognostic markers for ESCC. These plasma markers might facilitate clinical decision-making in the selection of prospective candidates who need meticulous follow-up for early detection of recurrences and additional treatments, such as neoadjuvant chemotherapy and postoperative chemotherapy in cases of ESCC. MiR-16, miR-21, miR-185, and miR-375 were found to be significantly higher in ESCC patients than in controls, suggesting that the four plasma miRNAs could potentially serve as novel diagnostic biomarkers for ESCC [60]. Moreover, the upregulated miR-16 and miR-21 in the plasma were correlated significantly to shortened progression-free survival and overall survival in ESCC patients, indicating that miR- 16 and miR-21 can predict poor survival in ESCC. The miR-20a and let-7a levels in plasma obviously change in ESCC patients, which could be used as a biomarker for the diagnosis of ESCC [61]. The serum expression of miR-218 is downregulated in EC patients and correlates with tumor differentiation, stage, and lymph node metastasis, so serum miR-218 may be a potential biomarker for early detection and clinical evaluation in patients with EC [62]. Compared with healthy controls, the miRNA-216a/b expression in plasma from ESCC patients is markedly decreased, and the miRNA-216a/b expression in plasma is inversely correlated with the TNM stage and lymph node metastasis [63]. The reduced miR-718 expression in plasma of ESCC patients may act as a potential diagnostic marker for ESCC [64]. The patients who have high miR-506 expression have a notably shorter survival time than those with low miR- 506 expression; thus, miRNA-506 can serve as a marker for the diagnosis and prognostic prediction of ESC [65].
MiRNAs and chemosensitivity of EC
Only a small number of studies have identified the significance of circulating miRNAs in blood as a predictive biomarker for chemoresistance in ESCC. Komatsu et al. [58,59] have demonstrated that a high plasma level of miR-21 indicates a high correlation with recurrence in ESCC patients. Also, He and his colleagues [66] further verified that the overexpression of miR-21 contributes to chemoresistance, and that circulating miR-21 could be a useful biomarker for predicting chemoresistance in ESCC patients. Sugimura et al. [67] found that let-7 expression in EC can predict the response to cisplatin-based chemotherapy and the modulated chemosensitivity role of let-7 to cisplatin by regulating the IL-6/STAT3 pathway. Convincing evidence has been offered by Chen et al. [56] that miR- 133a and miR-133b may predict chemosensitivity, which indicates their importance in being applied in the clinical treatment of ESCC. In order to provide new insights for ESCC treatment, Tian et al. [68] detected miR-218 in ESCC tissues and explored its effects on the biological features and chemosensitivity to cisplatin in the Eca109 cell. They found that, compared to the matched non-tumor tissues, miR-218 in the ESCC tissues was significantly increased. This supports the opinion that enforced expression of miR-218 significantly increases the sensitivity to CDDP in Eca109 cells.
Conclusion
The incidence rate of EC is showing rapid growth in the world. The treatment of EC is still routine surgery, radiotherapy, and chemotherapy, so the prognosis is poor. The 5-year survival rate of surgical patients is only 25%. It is encouraging to note that miRNAs have recently been shown to be involved in the whole the process of carcinogenesis, progression, and treatment of EC. Further exploration of the miRNA regulatory network will not only help us identify the pathogenesis of EC, but may also assist us in finding new therapies. However, further study is needed before miRNAs can be introduced into clinical application.
References
1. Yu, F., Fang, D., and Hu, J. (2012) MicroRNA and its roles in esophageal cancer. Medical Science Monitor International Medical Journal of Experimental & Clinical Research 18, RA22
2. Adachi, Y., Ohashi, H., Imsumran, A., Yamamoto, H., Matsunaga, Y., Taniguchi, H., Nosho, K., Suzuki, H., Sasaki, Y., and Arimura, Y. (2014) The effect of IGF-I receptor blockade for human esophageal squamous cell carcinoma and adenocarcinoma. Tumor Biology 35, 973-985
3. Meazza, CEnzinger, P. C., and Mayer, R. J. (2003) Esophageal cancer. New England Journal of Medicine 349, 2241-2252
4. Mei-Ling Zhu, J. H., MengYun Wang, Meng-Hong Sun, Li Jin, Xiaofeng Wang, Ya-Jun Yang, Jiu-Cun Wang, Leizhen Zheng, Jia-Qing Xiang, Qing-Yi Wei. (2014) Potentially functional polymorphisms in the ERCC2 gene and risk of Esophageal Squamous Cell Carcinoma in Chinese populations. Scientific Reports 4, 6281
5. Bartel, D. (2009) MicroRNAs: Target Recognition and Regulatory Functions. Cell 136, 215-233
6. Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843
7. Li, Z., and Rana, T. M. (2014) Therapeutic targeting of microRNAs: current status and future challenges. Nature Reviews Drug Discovery 13, 622
8. Gu, J., Wang, Y., and Wu, X. (2013) MicroRNA in the pathogenesis and prognosis of esophageal cancer.Current pharmaceutical design 19, 1292
9. Sharma, P., Saini, N., and Sharma, R. (2017) miR- 107 functions as a tumor suppressor in human esophageal squamous cell carcinoma and targets Cdc42. Oncology Reports 37, 3116-3127
10. Mao, Y., Liu, J., Zhang, D., and Li, B. (2016) miR-143 inhibits tumor progression by targeting FAM83F in esophageal squamous cell carcinoma. Tumor Biology 37, 9009-9022
11. Li, D., Shi, M., Ji, H., Chen, G., Jiang, H., and Wang, Z. (2016) MicroRNA-181d is a tumor suppressor in human esophageal squamous cell carcinoma inversely regulating Derlin-1. Oncology Reports 36, 2041-2048
12. Fu, M. G., Li, S., Yu, T. T., Qian, L. J., Cao, R. S., Zhu, H., Xiao, B., Jiao, C. H., Tang, N. N., and Ma, J. J. (2013) Differential expression of miR-195 in esophageal squamous cell carcinoma and miR-195 expression inhibits tumor cell proliferation and invasion by targeting of Cdc42. Febs Letters 587, 3471
13. Nie, Z. C., Weng, W. H., Shang, Y. S., Long, Y., Li, J., Xu, Y. T., and Li, Z. (2015) MicroRNA-126 is downregulated in human esophageal squamous cell carcinoma and inhibits the proliferation and migration in EC109 cell via PI3K/AKT signaling pathway. International Journal of Clinical & Experimental Pathology 8, 4745
14. Chen, H. X., Xu, X. X., Tan, B. Z., Zhang, Z., and Zhou, X. D. (2017) MicroRNA-29b Inhibits Angiogenesis by Targeting VEGFA through the MAPK/ERK and PI3K/Akt Signaling Pathways in Endometrial Carcinoma. Cellular Physiology & Biochemistry International Journal of Experimental Cellular Physiology Biochemistry & Pharmacology 41, 933
15. Zhu, L., Wang, Z., Fan, Q., Wang, R., and Sun, Y. (2014) microRNA-27a functions as a tumor s u p p re s s o r i n e s o p h a g e a l s q u a m o u s c e l l carcinoma by targeting KRAS. Oncology Reports 31, 280
16. Zhang, H. F., Alshareef, A., Wu, C., Jiao, J. W., Sorensen, P. H., Lai, R., Xu, L. Y., and Li, E. M. (2016) miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis 37, bgw079
17. Ding, D. P., Chen, Z. L., Zhao, X. H., Wang, J. W., Sun, J., Wang, Z., Tan, F. W., Tan, X. G., Li, B. Z., and Zhou, F. (2011) miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis 32, 1025
18. Zhang, K., Dai, L., Zhang, B., Xu, X., Shi, J., Fu, L., Chen, X., Li, J., and Bai, Y. (2015) miR-203 Is a Direct Transcriptional Target of E2F1 and Causes G1 Arrest in Esophageal Cancer Cells. Journal of cellular physiology 230, 903-910
19. Li, C., Zhou, X., Wang, Y., Jing, S., Yang, C., Sun, G., Liu, Q., Cheng, Y., and Wang, L. (2014) miR-210 regulates esophageal cancer cell proliferation by inducing G2/M phase cell cycle arrest through targeting PLK1. Molecular Medicine Reports 10, 2099
20. Mao, Y., Li, L., Liu, J., Wang, L., and Zhou, Y. (2016) MiR-495 inhibits esophageal squamous cell carcinoma progression by targeting Akt1. Oncotarget 7, 51223-51236
21. Liu, J., Mao, Y., Zhang, D., Hao, S., Zhang, Z., Li, Z., and Li, B. (2016) MiR-143 inhibits tumor cell proliferation and invasion by targeting STAT3 in esophageal squamous cell carcinoma. Cancer Letters 373, 97
22. Chen, Z., Zhao, L., Zhao, F., Yang, G., and Wang, J. (2016) MicroRNA-26b regulates cancer proliferation migration and cell cycle transition by suppressing TRAF5 in esophageal squamous cell carcinoma. Am J Transl Res 8, 1957-1970
23. Sun, N., Ye, L., Chang, T., Li, X., and Li, X. (2014) microRNA-195-Cdc42 axis acts as a prognostic factor of esophageal squamous cell carcinoma. International Journal of Clinical & Experimental Pathology 7, 6871-6879
24. Ma, J., Hong, L., Xu, G., Hao, J., Wang, R., Guo, H., Liu, J., Zhang, Y., Nie, Y., and Fan, D. (2016) miR-483-3p plays an oncogenic role in esophageal squamous cell carcinoma by targeting tumor suppressor EI24. Cell Biology International 40, 448
25. Wu, X. Z., Wang, K. P., Song, H. J., Xia, J. H., Jiang, Y., and Wang, Y. L. (2015) MiR-27a-3p promotes esophageal cancer cell proliferation via F-box and WD repeat domain-containing 7 (FBXW7) suppression. International Journal of Clinical & Experimental Medicine 8, 15556-15562
26. Zhu, Y., Xia, Y., Niu, H., and Chen, Y. (2014) MiR-16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cellular Physiology & Biochemistry International Journal of Experimental Cellular Physiology Biochemistry & Pharmacology 33, 1340
27. Li, H., Zheng, D., Zhang, B., Liu, L., Ou, J., Chen, W., Xiong, S., Gu, Y., and Yang, J. (2014) Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. Journal of Translational Medicine 12, 1-9
28. Yang, M., Liu, R., Li, X., Liao, J., Pu, Y., Pan, E., Yin, L., and Wang, Y. (2014) miRNA-183 suppresses apoptosis and promotes proliferation in esophageal cancer by targeting PDCD4. Molecules & Cells 37, 873-880
29. Zhang, W., Lei, C., Fan, J., and Wang, J. (2016) miR- 18a promotes cell proliferation of esophageal squamous cell carcinoma cells by increasing cylin D1 via regulating PTEN-PI3K-AKT-mTOR signaling axis. Biochemical & Biophysical Research Communications 477, 144-149
30. Yu, T., Cao, R., Li, S., Fu, M., Ren, L., Chen, W., Hong, Z., Qiang, Z., and Shi, R. (2015) MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer 15, 1-9
31. Lin, C., Liu, A., Zhu, J., Zhang, X., Wu, G., Ren, P., Wu, J., Li, M., Li, J., and Song, L. (2013) miR- 508 sustains phosphoinositide signalling and promotes aggressive phenotype of oesophageal squamous cell carcinoma. Nature Communications 5, 4620
32. Liu, F., Zheng, S., Liu, T., Liu, Q., Liang, M., Li, X., Sheyhidin, I., Lu, X., and Liu, W. (2013) MicroRNA-21 promotes the proliferation and inhibits apoptosis in Eca109 via activating ERK1/2/MAPK pathway. Molecular and Cellular Biochemistry 381, 115
33. Jing, Z., Wang, D., Shen, H., Liu, F., Jing, H., and Zhang, X. (2016) MicroRNA-153 inhibits tumor progression in esophageal squamous cell carcinoma by targeting SNAI1. Tumor Biology, 1-6
34. Song, Y., Li, J., Zhu, Y., Dai, Y., Zeng, T., Liu, L., Li, J., Wang, H., Qin, Y., and Zeng, M. (2014) MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget 5, 11669
35. Yanping, Siqi, Longbang, Chen, Haizhu, and Song. (2016) MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochimica Et Biophysica Sinica 48, 810-819
36. Matsushima, K., Isomoto, H., Yamaguchi, N., Inoue, N., Machida, H., Nakayama, T., Hayashi, T., Kunizaki, M., Hidaka, S., Nagayasu, T., Nakashima, M., Ujifuku, K., Mitsutake, N., Ohtsuru, A., Yamashita, S., Korpal, M., Kang, Y., Gregory, P. A., Goodall, G. J., Kohno, S., and Nakao, K. (2011) MiRNA-205 modulates cellular invasion and migration via regulating zinc finger E-box binding homeobox 2 expression in esophageal squamous cell carcinoma cells. J Transl Med 9, 30
37. Harazono, Y., Muramatsu, T., Endo, H., Uzawa, N., Kawano, T., Harada, K., Inazawa, J., and Kozaki, K. (2013) miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One 8, e62757
38. Li, S., Qin, X., Li, Y., Zhang, X., Niu, R., Zhang, H., Cui, A., An, W., and Wang, X. (2015) MiR- 133a suppresses the migration and invasion of esophageal cancer cells by targeting the EMT regulator SOX4. American Journal of Translational Research 7, 1390-1403
39. Wang, F., Wang, J., Yang, X., Chen, D., and Wang, L. (2016) MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagnostic Pathology 11, 88
40. Fang, D., Tao, L., Zheng, S., Liu, Q., Yang, C., Jian, Z., Chen, Y., Sheyhidin, I., and Lu, X. (2016) MiR- 106b promotes migration and invasion through enhancing EMT via downregulation of Smad 7 in Kazakh’s esophageal squamous cell carcinoma. Tumor Biology, 1-10
41. Han, Q., Zhang, H. Y., Zhong, B. L., Wang, X. J., Zhang, B., and Chen, H. (2016) MicroRNA-145 Inhibits Cell Migration and Invasion and Regulates Epithelial-Mesenchymal Transition (EMT) by Targeting Connective Tissue Growth Factor (CTGF) in Esophageal Squamous Cell Carcinoma. Medical Science Monitor International Medical Journal of Experimental & Clinical Research 22, 3925-3934
42. Cui, X. B., Su, L., Li, T. T., Hao, P., Jin, T. T., Zhang, S. M., Liu, C. X., Lan, Y., Shen, Y. Y., and Li, S. G. (2016) Targeting oncogenic PLCE1 by miR-145 impairs tumor proliferation and metastasis of esophageal squamous cell carcinoma. Oncotarget 7, 1777- 1795
43. Yan, S., Jiang, H., Fang, S., Yin, F., Wang, Z., Jia, Y., Sun, X., Wu, S., Jiang, T., and Mao, A. (2015) MicroRNA-340 Inhibits Esophageal Cancer Cell Growth and Invasion by Targeting Phosphoserine Aminotransferase 1. Cellular Physiology & Biochemistry International Journal of Experimental Cellular Physiology Biochemistry & Pharmacology 37, 375-386
44. Mei, L. L., Qiu, Y. T., Zhang, B., and Shi, Z. Z. (2017) MicroRNAs in esophageal squamous cell carcinoma: Potential biomarkers and therapeutic targets. Cancer Biomarkers 19, 1
45. Wang, Y., Zhang, J., Zhao, W., Wang, D., Ma, W., Shang, S., Feng, C., and Yu, H. (2017) MicroRNA expression in esophageal squamous cell carcinoma: Novel diagnostic and prognostic biomarkers. Molecular Medicine Reports 15, 3833-3839
46. Li, J., Li, M., Gao, F., and Ge, X. (2016) Serum microRNA-15a level acts as a potential diagnostic and prognostic biomarker for human esophageal squamous cell carcinoma. Cancer Biomarkers 18, 1-7
47. Hara, K., Miyazaki, T., Yokobori, T., Yoshida, T., Kumakura, Y., Honjyo, H., Sakai, M., Sohda, M.,Fukuchi, M., and Kuwano, H. (2017) Significance and Function of MicroRNA-7 in Oesophageal Squamous Cell Carcinoma. Anticancer Res 37, 1043-1048
48. Li, B., Xu, W. W., Han, L., Chan, K. T., Tsao, S. W., Lee, N. P., Law, S., Xu, L. Y., Li, E. M., and Chan, K. W. (2017) MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene 36, 3986
49. Tomoyuki, O., Hirohumi, K., Takeshi, M., Shinichi, S., Isaya, H., Shozo, H., Takuya, N., and Yutaka, S. (2016) The expression of microRNA 574-3p as a predictor of postoperative outcome in patients with esophageal squamous cell carcinoma. World Journal of Surgical Oncology 14, 228
50. Wang, C., Guan, S., Liu, F., Chen, X., Han, L., Wang, D., Nesa, E. U., Wang, X., Bao, C., and Wang, N. (2016) Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. British Journal of Cancer 114, 290
51. Zhou, S., Yang, B., Zhao, Y., Xu, S., Zhang, H., and Li, Z. (2014) Prognostic value of microRNA-100 in esophageal squamous cell carcinoma. Journal of Surgical Research 192, 515-520
52. Xu, X. L., Jiang, Y. H., Feng, J. G., Su, D., Chen, P. C., and Mao, W. M. (2014) MicroRNA-17, microRNA- 18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Annals of Thoracic Surgery 97, 1037-1045
53. Hirajima, S., Komatsu, S., Ichikawa, D., Takeshita, H., Konishi, H., Shiozaki, A., Morimura, R., Tsujiura, M., Nagata, H., and Kawaguchi, T. (2013) Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. British Journal of Cancer 108, 1822
54. Zhao, Y., Schetter, A. J., Yang, G. B., Nguyen, G., Mathé, E. A., Li, P., Cai, H., Yu, L., Liu, F., and Hang, D. (2013) microRNA and inflammatory gene expression as prognostic marker for overall survival in esophageal squamous cell carcinoma. International journal of cancer 132, 2901–2909
55. Tanaka, K., Miyata, H., Yamasaki, M., Sugimura, K., Takahashi, T., Kurokawa, Y., Nakajima, K., Takiguchi, S., Mori, M., and Doki, Y. (2013) Circulating miR-200c Levels Significantly Predict Response to Chemotherapy and Prognosis of Patients Undergoing Neoadjuvant Chemotherapy for Esophageal Cancer. Annals of Surgical Oncology 20, 607-615
56. Chen, G., Peng, J., Zhu, W., Tao, G., Song, Y., Zhou, X., and Wang, W. (2014) Combined downregulation of microRNA-133a and microRNA-133b predicts chemosensitivity of patients with esophageal squamous cell carcinoma undergoing paclitaxel based chemotherapy. Medical Oncology 31, 1-8
57. Liu, R., Liao, J., Yang, M., Shi, Y., Peng, Y., Wang, Y., Pan, E., Guo, W., Pu, Y., and Yin, L. (2012) Circulating miR-155 expression in plasma: a potential biomarker for early diagnosis of esophageal cancer in humans. Journal of Toxicology & Environmental Health Part A 75, 1154
58. Komatsu, S., Ichikawa, D., Takeshita, H., Tsujiura, M., Morimura, R., Nagata, H., Kosuga, T., Iitaka, D., Konishi, H., and Shiozaki, A. (2011) Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. British Journal of Cancer 105, 104-111
59. Komatsu, S., Ichikawa, D., Takeshita, H., Konishi, H., Nagata, H., Hirajima, S., Kawaguchi, T., Arita, T., Shiozaki, A., and Fujiwara, H. (2012) Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opinion on Biological Therapy 1, 53-59
60. Li, B. X., Yu, Q., Shi, Z. L., Li, P., and Fu, S. (2014) Circulating microRNAs in esophageal squamous cell carcinoma: association with locoregional staging and survival. International Journal of Clinical & Experimental Medicine 8, 7241
61. He, F. C., Meng, W. W., Qu, Y. H., Zhou, M. X., He, J., Lv, P., and Ming, L. (2015) Expression of circulating microRNA-20a and let-7a in esophageal squamous cell carcinoma. World Journal of Gastroenterology 21, 4660-4665
62. Jiang, Z., Song, Q., Yang, S., Zeng, R., Li, X., Jiang, C., Ding, W., Zhang, J., and Zheng, Y. (2015) Serum microRNA-218 is a potential biomarker for esophageal cancer. Cancer Biomarkers 15, 381
63. Dong, S., Yin, H., Dong, C., Sun, K., Lv, P., Meng, W., Ming, L., and He, F. (2016) Predictive Value of Plasma MicroRNA-216a/b in the Diagnosis of Esophageal Squamous Cell Carcinoma. Disease Markers,2016,(2016-2-18) 2016, 1-8
64. Sun, L., Dong, S., Dong, C., Sun, K., Meng, W., Lv, P., Yin, H., Ming, L., and He, F. (2016) Predictive value of plasma miRNA-718 for esophageal squamous cell carcinoma. Cancer Biomarkers 16, 265
65. Li, S. P., Su, H. X., Zhao, D., and Guan, Q. L. (2016) Plasma miRNA-506 as a Prognostic Biomarker for Esophageal Squamous Cell Carcinoma. Medical Science Monitor International Medical Journal of Experimental & Clinical Research 22, 2195
66. Komatsu, S., Ichikawa, D., Kawaguchi, T., Miyamae, M., Okajima, W., Ohashi, T., Imamura, T., Kiuchi, J., Konishi, H., and Shiozaki, A. (2016) Circulating miR-21 as an independent predictive biomarker for chemoresistance in esophageal squamous cell carcinoma. Am J Cancer Res 6, 1511
67. Sugimura, K., Miyata, H., Tanaka, K., Hamano, R., Takahashi, T., Kurokawa, Y., Yamasaki, M., Nakajima, K., Takiguchi, S., and Mori, M. (2012) Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clinical Cancer Research An Official Journal of the American Association for Cancer Research 18, 5144-5153
68. Tian, H., Hou, L., Xiong, Y. M., Huang, J. X., She, Y. J., Bi, X. B., and Song, X. R. (2015) miR-218 suppresses tumor growth and enhances the chemosensitivity of esophageal squamous cell carcinoma to cisplatin. Oncology Reports 33, 981